
Co-Director LSHTM AMR Centre @lshtmamrcentre.bsky.social
holtlab.net | klebnet.org | typhoidgenomics.org | amr.lshtm.ac.uk
Most are not on bluesky (yet!)…

Most are not on bluesky (yet!)…


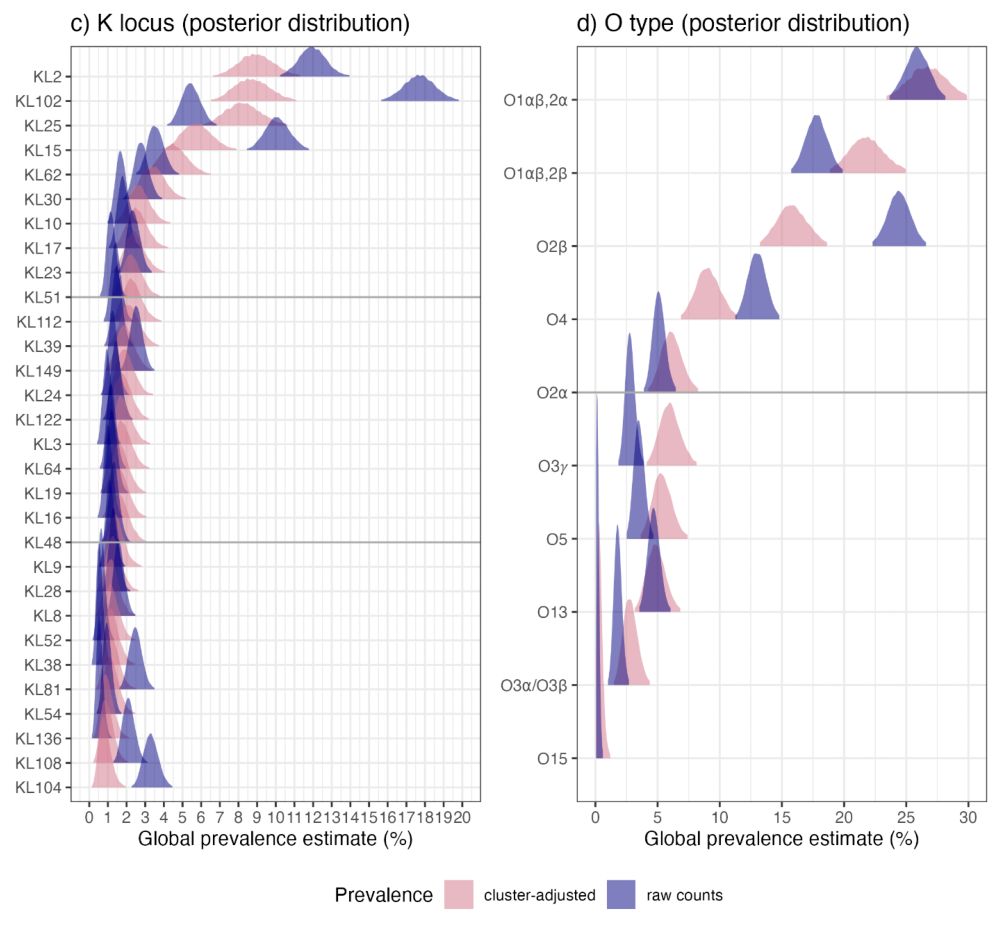
Yes this is a lot of antigens! But it has been done for Streptococcus pneumoniae.

Yes this is a lot of antigens! But it has been done for Streptococcus pneumoniae.



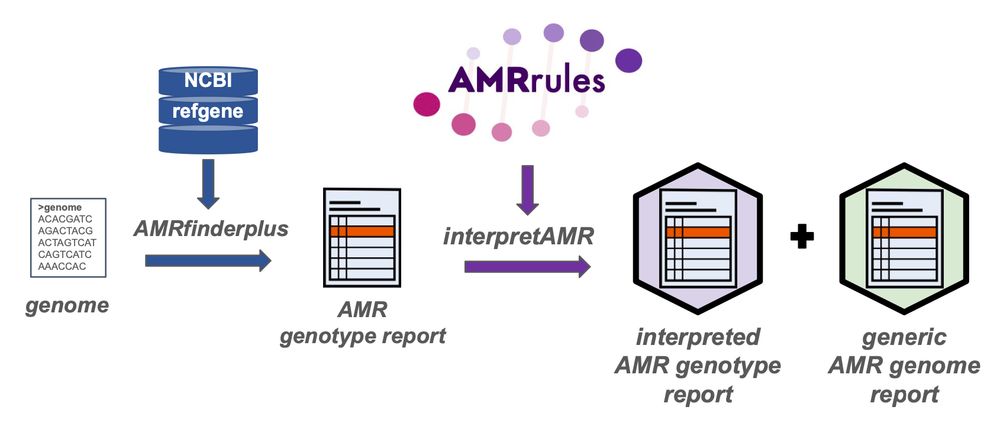
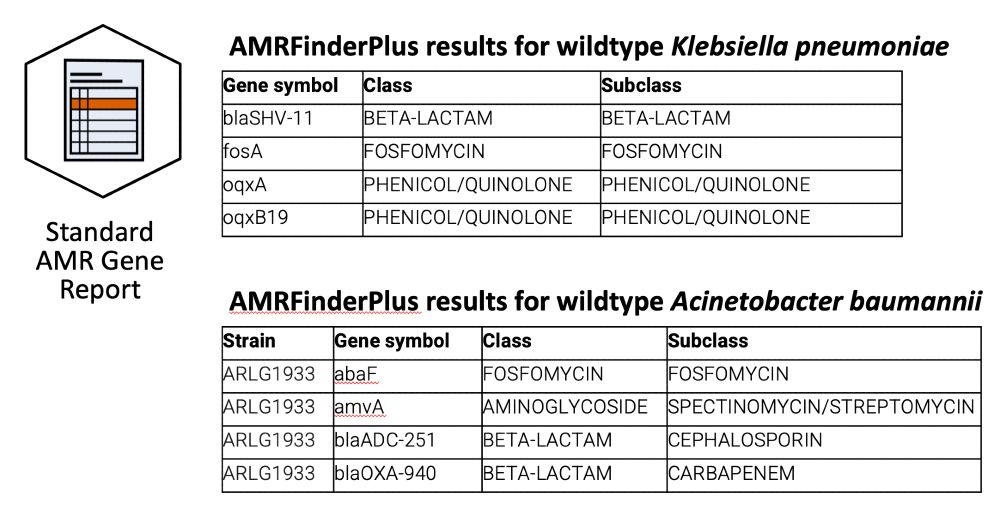
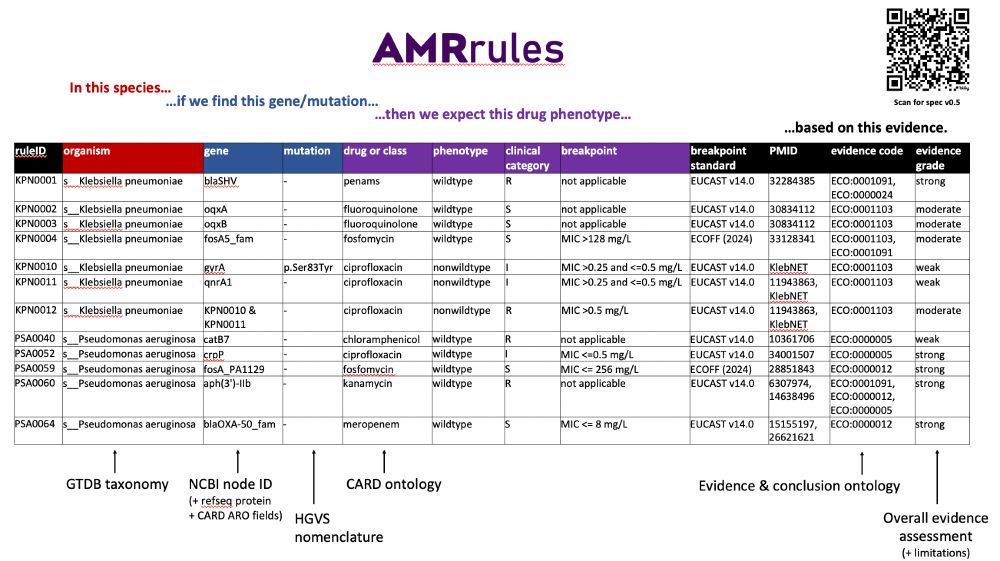
interpretamr.github.io/AMRrules

interpretamr.github.io/AMRrules
#ABPHM25
www.biorxiv.org/content/10.1...

#ABPHM25
www.biorxiv.org/content/10.1...




pubmed.ncbi.nlm.nih.gov/27067320/

pubmed.ncbi.nlm.nih.gov/27067320/
pubmed.ncbi.nlm.nih.gov/26100894/

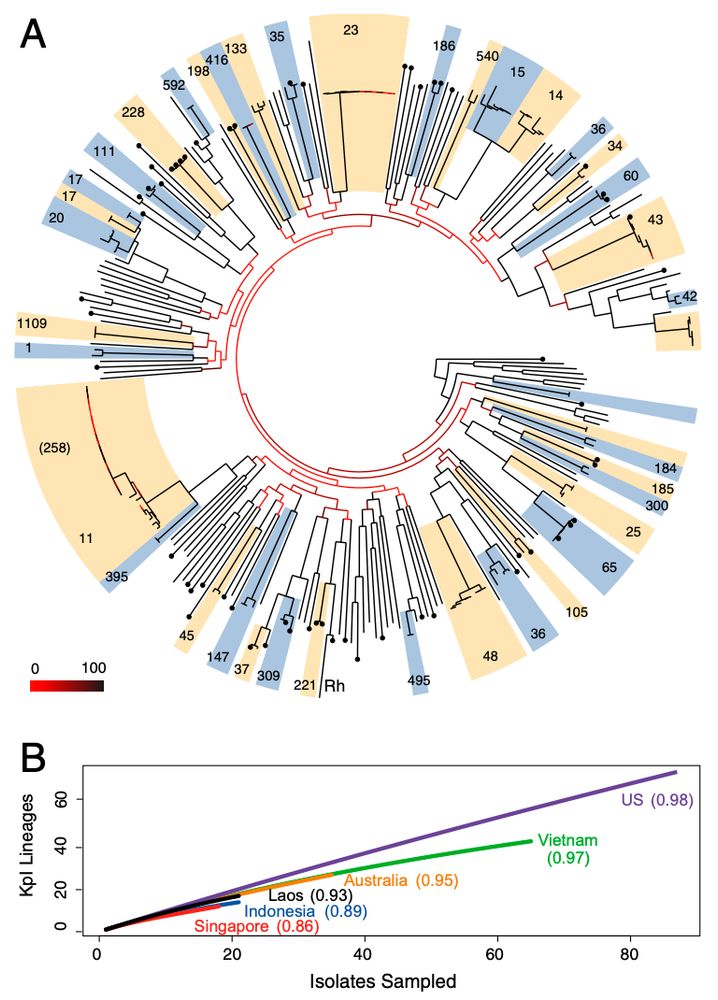
pubmed.ncbi.nlm.nih.gov/26100894/
pubmed.ncbi.nlm.nih.gov/24961694/
…and the introduction of core-genome MLST for Klebsiella
pubmed.ncbi.nlm.nih.gov/25341126/

pubmed.ncbi.nlm.nih.gov/24961694/
…and the introduction of core-genome MLST for Klebsiella
pubmed.ncbi.nlm.nih.gov/25341126/
pubmed.ncbi.nlm.nih.gov/22914622/
…followed closely by the first study tracking KPC plasmid transfer in the hospital (2014)
pubmed.ncbi.nlm.nih.gov/25232178/

pubmed.ncbi.nlm.nih.gov/22914622/
…followed closely by the first study tracking KPC plasmid transfer in the hospital (2014)
pubmed.ncbi.nlm.nih.gov/25232178/
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/
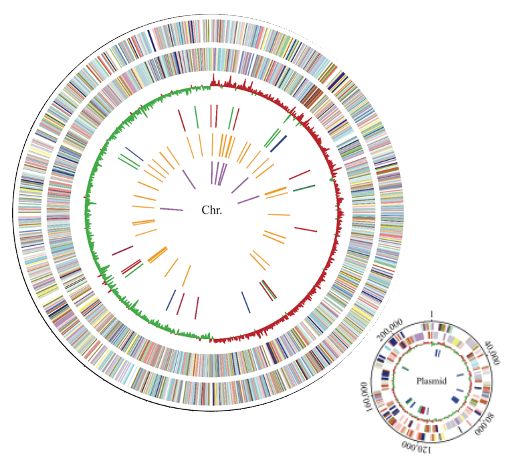
2009 was a big year for Klebs!
- First genome published
- First description of ST258 KPC-producing clone
pubmed.ncbi.nlm.nih.gov/19218573/
- First description of NDM-1 beta-lactamase
pubmed.ncbi.nlm.nih.gov/19770275/

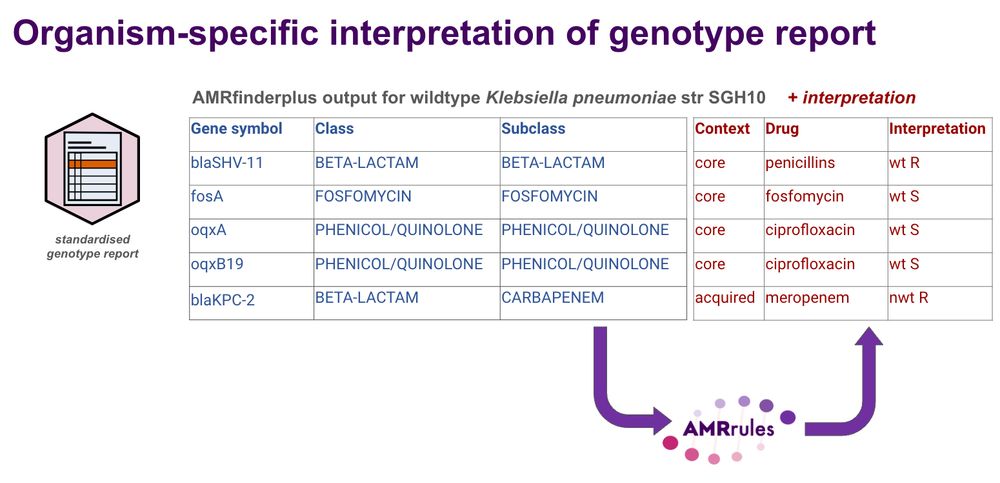
We need to do the same for AMR, to dig below species level and understand which variants are high-risk for transmission 🧫🧬

We need to do the same for AMR, to dig below species level and understand which variants are high-risk for transmission 🧫🧬

