🪄 The real power comes from combining 𝐭𝐲𝐏𝐏𝐢𝐧𝐠 with other methods (𝐯𝐂𝐨𝐧𝐓𝐀𝐂𝐓, 𝐌𝐌-𝐆𝐑𝐂, 𝐠𝐞𝐍𝐨𝐦𝐚𝐝) → also enables systematic discovery of 𝐡𝐢𝐠𝐡𝐥𝐲 𝐝𝐢𝐯𝐞𝐫𝐬𝐞 𝐚𝐧𝐝 𝐧𝐨𝐯𝐞𝐥 𝐭𝐲𝐩𝐞𝐬 𝐨𝐟 𝐏-𝐏𝐬.
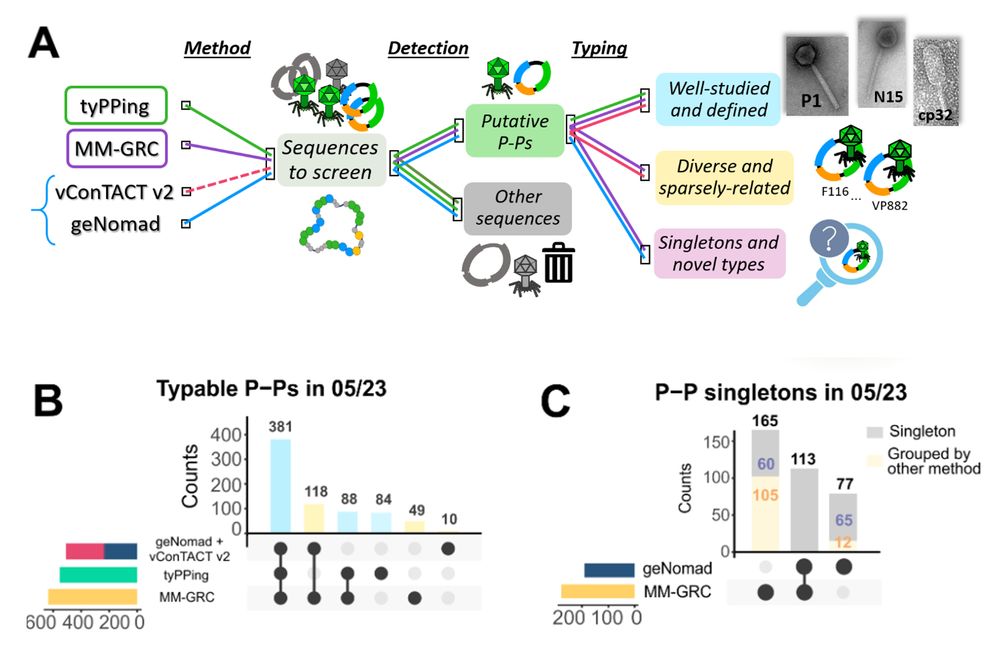
🪄 The real power comes from combining 𝐭𝐲𝐏𝐏𝐢𝐧𝐠 with other methods (𝐯𝐂𝐨𝐧𝐓𝐀𝐂𝐓, 𝐌𝐌-𝐆𝐑𝐂, 𝐠𝐞𝐍𝐨𝐦𝐚𝐝) → also enables systematic discovery of 𝐡𝐢𝐠𝐡𝐥𝐲 𝐝𝐢𝐯𝐞𝐫𝐬𝐞 𝐚𝐧𝐝 𝐧𝐨𝐯𝐞𝐥 𝐭𝐲𝐩𝐞𝐬 𝐨𝐟 𝐏-𝐏𝐬.
👉 Applied to 𝐜𝐨𝐦𝐩𝐥𝐞𝐭𝐞 + 𝐝𝐫𝐚𝐟𝐭 bacterial genomes → many new high-confidence P-Ps 🎯
👉 Large plasmid dataset (>38k plasmids) processed in just ~8 minutes ⏱️
👉 New P-Ps detected in draft genomes of carbapenem-resistant strains→ confirmed as active by experiments 🧪

👉 Applied to 𝐜𝐨𝐦𝐩𝐥𝐞𝐭𝐞 + 𝐝𝐫𝐚𝐟𝐭 bacterial genomes → many new high-confidence P-Ps 🎯
👉 Large plasmid dataset (>38k plasmids) processed in just ~8 minutes ⏱️
👉 New P-Ps detected in draft genomes of carbapenem-resistant strains→ confirmed as active by experiments 🧪
1️⃣ Compare proteins to 763 signature profiles
2️⃣ Process via two analytical branches
3️⃣ Assign 𝐜𝐨𝐧𝐟𝐢𝐝𝐞𝐧𝐜𝐞 𝐥𝐞𝐯𝐞𝐥𝐬 (High / Medium / Low) for simple interpretation.
✨ Currently covers the most prevalent groups (with experimentally proven members).
🚀 𝐍𝐞𝐰 𝐠𝐫𝐨𝐮𝐩𝐬 𝐜𝐚𝐧 & 𝐰𝐢𝐥𝐥 𝐛𝐞 𝐚𝐝𝐝𝐞𝐝!

1️⃣ Compare proteins to 763 signature profiles
2️⃣ Process via two analytical branches
3️⃣ Assign 𝐜𝐨𝐧𝐟𝐢𝐝𝐞𝐧𝐜𝐞 𝐥𝐞𝐯𝐞𝐥𝐬 (High / Medium / Low) for simple interpretation.
✨ Currently covers the most prevalent groups (with experimentally proven members).
🚀 𝐍𝐞𝐰 𝐠𝐫𝐨𝐮𝐩𝐬 𝐜𝐚𝐧 & 𝐰𝐢𝐥𝐥 𝐛𝐞 𝐚𝐝𝐝𝐞𝐝!


