Justus Kebschull
@justuskebschull.bsky.social
Neuroscientist of sorts. Head of the Kebschull Lab at Johns Hopkins BME. https://www.kebschull-lab.org/
Want to learn everything there is to know about barcoded connectomics, brain clearing, and modern neuroanatomy? There is still time to apply for the CSHL High Throughput Neuroanatomy course! Stipends are available. meetings.cshl.edu/courses.aspx...

July 19, 2025 at 10:39 PM
Want to learn everything there is to know about barcoded connectomics, brain clearing, and modern neuroanatomy? There is still time to apply for the CSHL High Throughput Neuroanatomy course! Stipends are available. meetings.cshl.edu/courses.aspx...
And even when neurons innervate the same kind of regions, they can do so in different ways. VTA and SNc DA neurons projecting to the dorsal striatum project fundamentally differently. SNc neurons innervate more subregions and broadcast randomly to these regions, while VTA neurons are highly specific
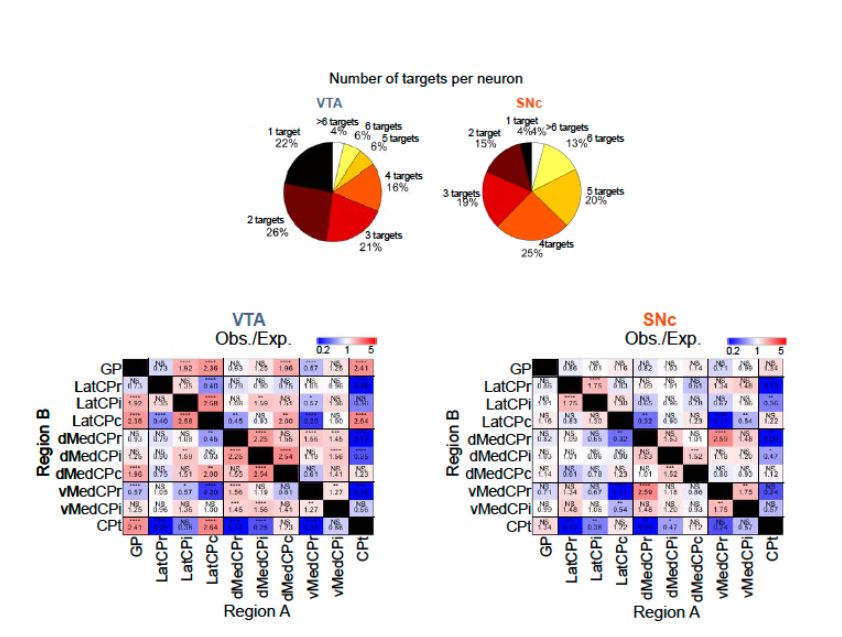
July 7, 2025 at 3:13 PM
And even when neurons innervate the same kind of regions, they can do so in different ways. VTA and SNc DA neurons projecting to the dorsal striatum project fundamentally differently. SNc neurons innervate more subregions and broadcast randomly to these regions, while VTA neurons are highly specific
This large number of clusters is not an effect of splitting a stochastically wired system. Co-innervation patterns and projection motifs are often far from the null model of independent projection choice.


July 7, 2025 at 3:13 PM
This large number of clusters is not an effect of splitting a stochastically wired system. Co-innervation patterns and projection motifs are often far from the null model of independent projection choice.
We find over 30 connectomic cell types, way more than would have been expected from transcriptomic cell types. Overall, our data agrees nicely with what we know, but boy, is there more structure to this system. Maybe easier to see from the dot plots.

July 7, 2025 at 3:13 PM
We find over 30 connectomic cell types, way more than would have been expected from transcriptomic cell types. Overall, our data agrees nicely with what we know, but boy, is there more structure to this system. Maybe easier to see from the dot plots.
To showcase POINTseq's utility, we then applied it to the midbrain dopaminergic system, mapping >3,800 DA neurons in the VTA and SNc.


July 7, 2025 at 3:13 PM
To showcase POINTseq's utility, we then applied it to the midbrain dopaminergic system, mapping >3,800 DA neurons in the VTA and SNc.
We validated POINTseq in the mouse motor cortex using two cre lines and AAVretro-cre to define cell types of interest.

July 7, 2025 at 3:13 PM
We validated POINTseq in the mouse motor cortex using two cre lines and AAVretro-cre to define cell types of interest.
This works all over the brain

July 7, 2025 at 3:13 PM
This works all over the brain
POINTseq achieves cell type specific barcoding by pseudotyping barcoded Sindbis virus with the structural proteins of Alphavirus CHIKV, and overexpressing CHIKV cellular receptor Mxra8 in target cells. Mxra8 isn't normally expressed in mouse neurons, enabling cell type specific infection.

July 7, 2025 at 3:13 PM
POINTseq achieves cell type specific barcoding by pseudotyping barcoded Sindbis virus with the structural proteins of Alphavirus CHIKV, and overexpressing CHIKV cellular receptor Mxra8 in target cells. Mxra8 isn't normally expressed in mouse neurons, enabling cell type specific infection.
Finally have time to add some more meat here. MAPseq2 is what it says on the tin. Better and cheaper. Use this going forward for anything involving MAPseq, including BARseq, BRICseq, and ConnectID!

July 7, 2025 at 2:53 PM
Finally have time to add some more meat here. MAPseq2 is what it says on the tin. Better and cheaper. Use this going forward for anything involving MAPseq, including BARseq, BRICseq, and ConnectID!
The Kebschull and @rezakalhor.bsky.social labs are recruiting a joint postdoc for an exciting project at the interface of phylodynamics and neuroscience. Come join us at JHU BME. Spread the word, and contact us if you are interested. #hiring 🧪🧠. More on www.kebschull-lab.org and below.

November 18, 2024 at 1:01 PM
The Kebschull and @rezakalhor.bsky.social labs are recruiting a joint postdoc for an exciting project at the interface of phylodynamics and neuroscience. Come join us at JHU BME. Spread the word, and contact us if you are interested. #hiring 🧪🧠. More on www.kebschull-lab.org and below.
Hello there! We are the Kebschull lab at Johns Hopkins University and love brains, barcodes, viruses, and sequencing. We develop new technology and work on a zoo of animals to understand how brains change over evolution and an animal's lifetime. Check out our lab at www.kebschull-lab.org 🧠🧪

November 15, 2024 at 2:06 PM
Hello there! We are the Kebschull lab at Johns Hopkins University and love brains, barcodes, viruses, and sequencing. We develop new technology and work on a zoo of animals to understand how brains change over evolution and an animal's lifetime. Check out our lab at www.kebschull-lab.org 🧠🧪

