JungmannLab
@jungmannlab.bsky.social
Our group at LMU Munich and @mpibiochem.bsky.social uses DNA nanotechnology to develop next-generation super-resolution microscopy techniques. #DNAPAINT
Thanks to all who made this possible! @eduardunterauer.bsky.social @evaschentarra.bsky.social @ipachmayr.bsky.social @taishatashrin.bsky.social Jisoo Kwon Sebastian Strauss, @jekristina.bsky.social @rafalkowalew.bsky.social @opazo.bsky.social @forna.bsky.social @lumasullo.bsky.social (6/6)
October 2, 2025 at 11:37 AM
Thanks to all who made this possible! @eduardunterauer.bsky.social @evaschentarra.bsky.social @ipachmayr.bsky.social @taishatashrin.bsky.social Jisoo Kwon Sebastian Strauss, @jekristina.bsky.social @rafalkowalew.bsky.social @opazo.bsky.social @forna.bsky.social @lumasullo.bsky.social (6/6)
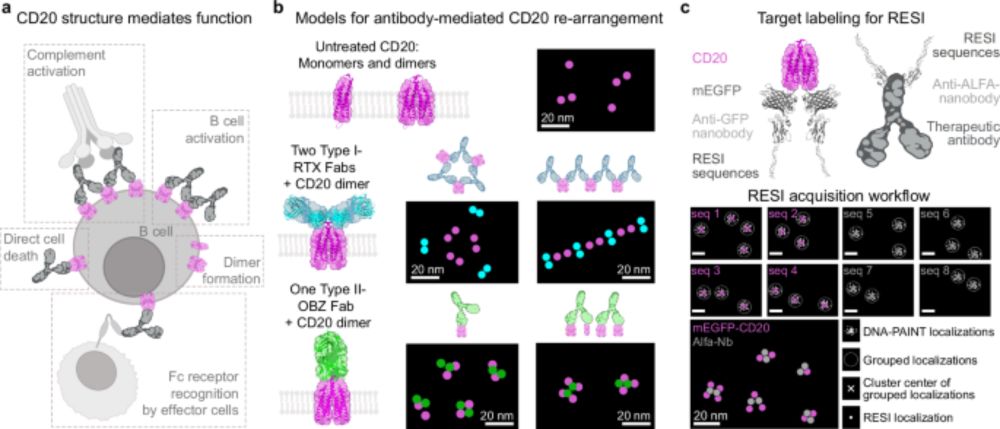
Within this neuronal atlas we can reveal the three synapse classes, excitatory, inhibitory and the recently discovered mixed synapse. Organelle imaging of Peroxisomes (Pmp70) and the Golgi Apparatus (Golga5) reveals rare contact sides and even fused particles. (5/6)

October 2, 2025 at 11:37 AM
Within this neuronal atlas we can reveal the three synapse classes, excitatory, inhibitory and the recently discovered mixed synapse. Organelle imaging of Peroxisomes (Pmp70) and the Golgi Apparatus (Golga5) reveals rare contact sides and even fused particles. (5/6)
To show the power of the technique, we acquired a 13-plex 200 x 200 µm2 neuronal atlas in 3D. With this atlas we map the interaction architecture of three neurons, resolving organelles, cytoskeleton, vesicles and synapses at single-protein resolution. (4/6)

October 2, 2025 at 11:37 AM
To show the power of the technique, we acquired a 13-plex 200 x 200 µm2 neuronal atlas in 3D. With this atlas we map the interaction architecture of three neurons, resolving organelles, cytoskeleton, vesicles and synapses at single-protein resolution. (4/6)
We demonstrate speed-optimized left-handed DNA-PAINT by characterizing the sequence binding kinetics and resolving three main microscopy benchmarking targets, mitochondria, microtubules and nuclear pore complexes with <5 nm localization precision. (3/6)

October 2, 2025 at 11:37 AM
We demonstrate speed-optimized left-handed DNA-PAINT by characterizing the sequence binding kinetics and resolving three main microscopy benchmarking targets, mitochondria, microtubules and nuclear pore complexes with <5 nm localization precision. (3/6)
The mirrored design of left-handed oligonucleotides allows the extension of the common 6 speed-sequences R1-R6 with their analogs L1-L6, enabling 12 target multiplexing with a standard secondary label-free DNA-PAINT workflow. (2/6)

October 2, 2025 at 11:37 AM
The mirrored design of left-handed oligonucleotides allows the extension of the common 6 speed-sequences R1-R6 with their analogs L1-L6, enabling 12 target multiplexing with a standard secondary label-free DNA-PAINT workflow. (2/6)
Congratulations to everyone involved: @ipachmayr.bsky.social, @lumasullo.bsky.social, @susannereinhardt.bsky.social, Jisoo Kwon, Ondřej Skořepa, Maite Llop, Sylvia Herter, Marina Bacac and Christian Klein. (6/6)
Read the full story here: www.nature.com/articles/s41...
Read the full story here: www.nature.com/articles/s41...
Resolving the structural basis of therapeutic antibody function in cancer immunotherapy with RESI - Nature Communications
The nanoscale organization of the antigen-antibody complexes influences the therapeutic action of monoclonal antibodies. Here, the authors present a multi-target 3D RESI imaging assay for the nanomete...
www.nature.com
July 28, 2025 at 8:36 AM
Congratulations to everyone involved: @ipachmayr.bsky.social, @lumasullo.bsky.social, @susannereinhardt.bsky.social, Jisoo Kwon, Ondřej Skořepa, Maite Llop, Sylvia Herter, Marina Bacac and Christian Klein. (6/6)
Read the full story here: www.nature.com/articles/s41...
Read the full story here: www.nature.com/articles/s41...
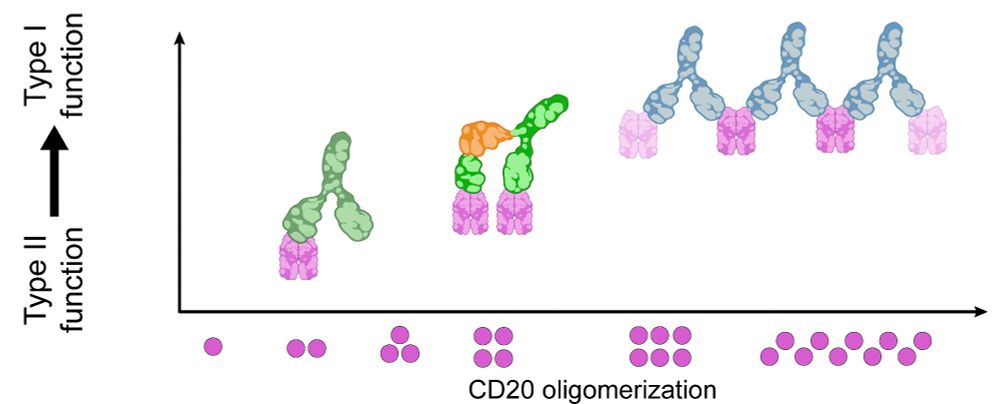
The shift from Type II to Type I function reveals a structure–function continuum for anti-CD20 antibodies, showing that receptor arrangements dictate mechanism of action. RESI provides a platform for structure-guided antibody development, applicable far beyond CD20. (5/6)
July 28, 2025 at 8:36 AM
The shift from Type II to Type I function reveals a structure–function continuum for anti-CD20 antibodies, showing that receptor arrangements dictate mechanism of action. RESI provides a platform for structure-guided antibody development, applicable far beyond CD20. (5/6)
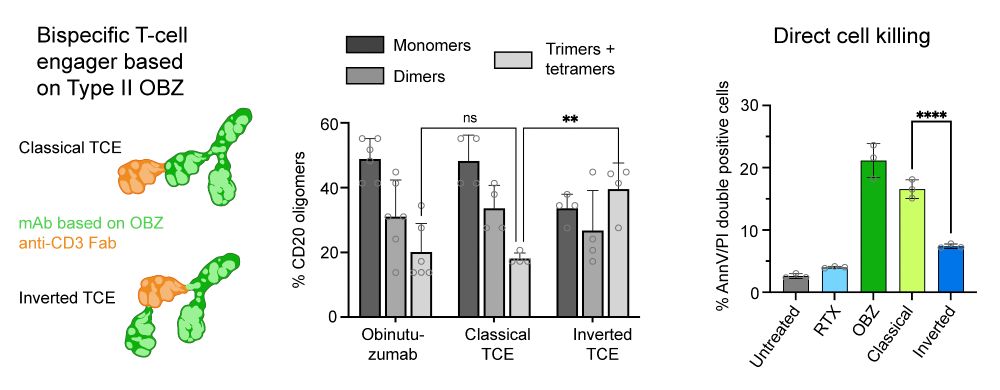
We showed a direct link between CD20 oligomerization and function by investigating OBZ-based T-cell engagers (TCEs). An increased IgG flexibility in the 2+1 TCE format lead to increased CD20 tetramerization, without higher-order clustering, resulting in a reduction of direct cytotoxicity. (4/6)
July 28, 2025 at 8:36 AM
We showed a direct link between CD20 oligomerization and function by investigating OBZ-based T-cell engagers (TCEs). An increased IgG flexibility in the 2+1 TCE format lead to increased CD20 tetramerization, without higher-order clustering, resulting in a reduction of direct cytotoxicity. (4/6)
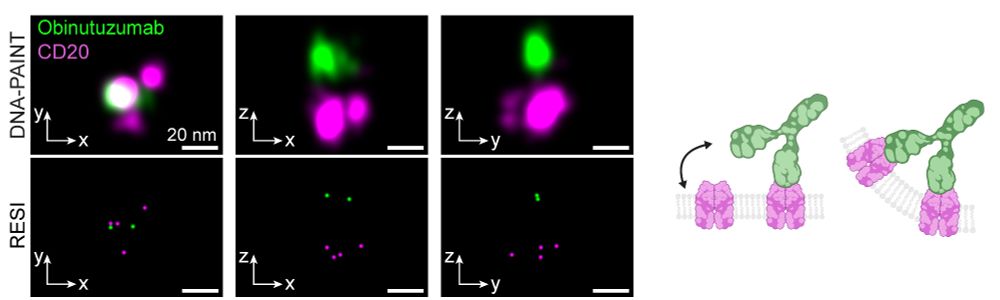
In contrast, Type II antibodies like Obinutuzumab and H299 induced limited oligomerization to dimers, trimers and tetramers, consistent with their role in promoting direct tumor cell death rather than complement activation. (3/6)
July 28, 2025 at 8:36 AM
In contrast, Type II antibodies like Obinutuzumab and H299 induced limited oligomerization to dimers, trimers and tetramers, consistent with their role in promoting direct tumor cell death rather than complement activation. (3/6)
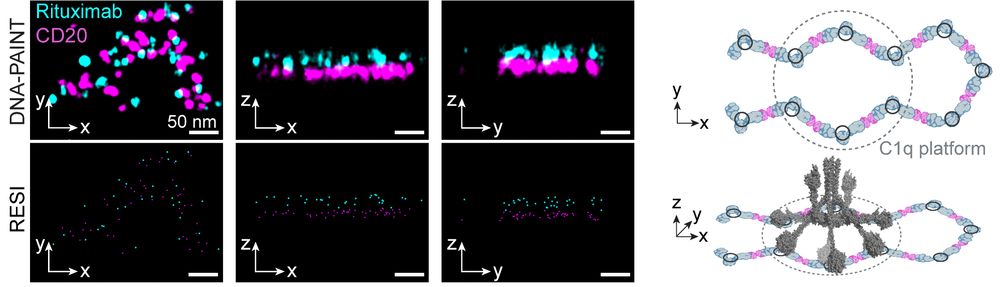
By imaging intact cells, we could see these therapeutic antibodies in action: Type I antibodies like Rituximab and Ofatumumab formed extended chains of CD20 hexamers or larger, creating platforms compatible with complement protein binding, mediating cancer cell killing. (2/6)
July 28, 2025 at 8:36 AM
By imaging intact cells, we could see these therapeutic antibodies in action: Type I antibodies like Rituximab and Ofatumumab formed extended chains of CD20 hexamers or larger, creating platforms compatible with complement protein binding, mediating cancer cell killing. (2/6)
May 7, 2025 at 3:21 PM
@moniquehonsa.bsky.social , @philippsteen.bsky.social , Larissa Heinze, Shuhan Xu, Heinrich Grabmayr, Isabelle Pachmayr, Susanne C. M. Reinhardt, Ana Perovic, Jisoo Kwon, Ethan P. Oxley, Ross A. Dickins, Maartje M. C. Bastings, Ian A. Parish
May 7, 2025 at 2:43 PM
@moniquehonsa.bsky.social , @philippsteen.bsky.social , Larissa Heinze, Shuhan Xu, Heinrich Grabmayr, Isabelle Pachmayr, Susanne C. M. Reinhardt, Ana Perovic, Jisoo Kwon, Ethan P. Oxley, Ross A. Dickins, Maartje M. C. Bastings, Ian A. Parish
Big congrats to @lumasullo.bsky.social and @rafalkowalew.bsky.social who led the project as well as other co-authors that contributed to this work!!
May 7, 2025 at 2:43 PM
Big congrats to @lumasullo.bsky.social and @rafalkowalew.bsky.social who led the project as well as other co-authors that contributed to this work!!
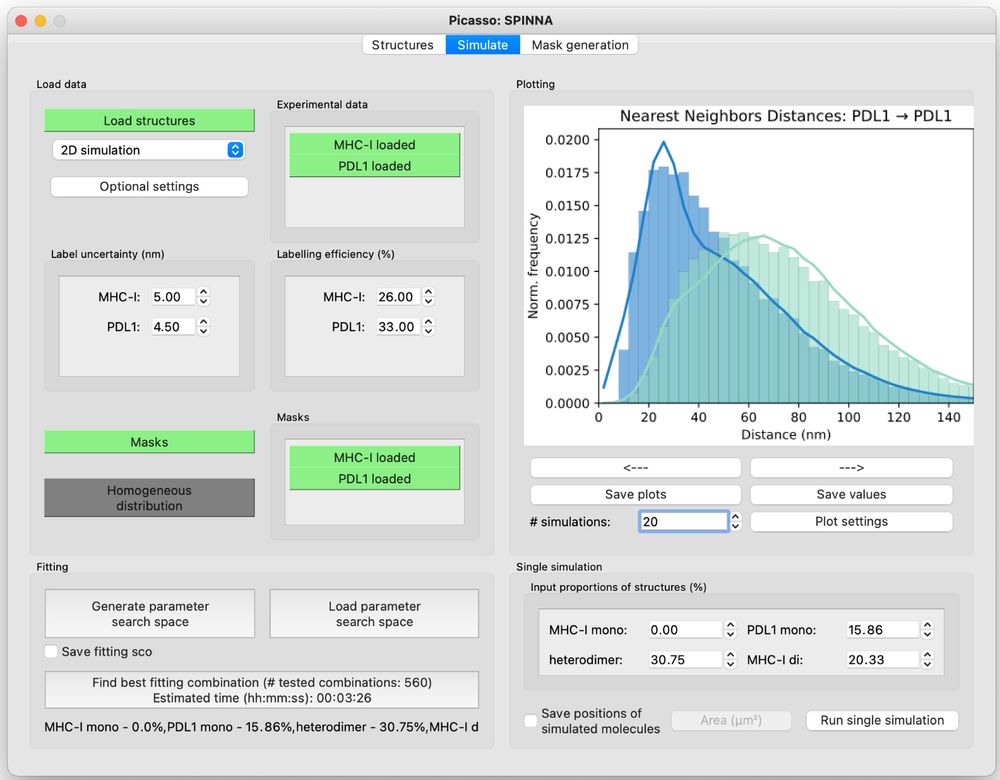
To facilitate SPINNA’s widespread use in the scientific community, we offer an open-source Python implementation and a GUI available in the latest version of Picasso (github.com/jungmannlab/..., picassosr.readthedocs.io/en/latest/sp...). 7/7
May 7, 2025 at 2:43 PM
To facilitate SPINNA’s widespread use in the scientific community, we offer an open-source Python implementation and a GUI available in the latest version of Picasso (github.com/jungmannlab/..., picassosr.readthedocs.io/en/latest/sp...). 7/7
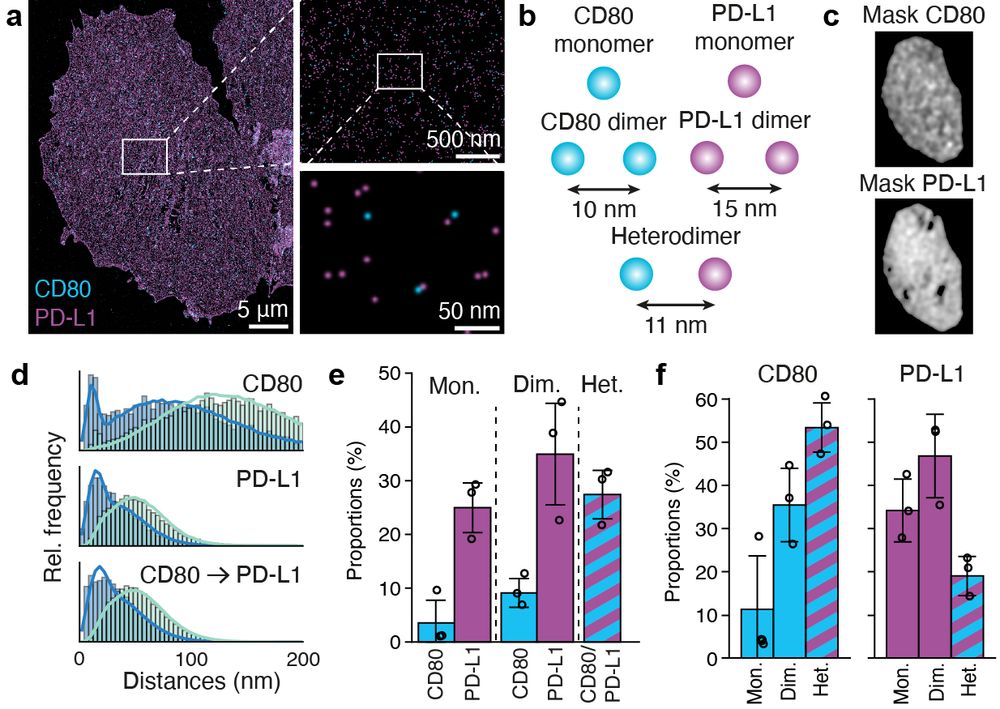
Finally, we investigate the dimerization of CD80 and PD-L1, key surface ligands involved in immune cell signaling. 6/7
May 7, 2025 at 2:43 PM
Finally, we investigate the dimerization of CD80 and PD-L1, key surface ligands involved in immune cell signaling. 6/7
We further quantitatively evaluate the oligomerization of the Epidermal Growth Factor Receptor (EGFR) upon binding of its ligand, EGF. 5/7
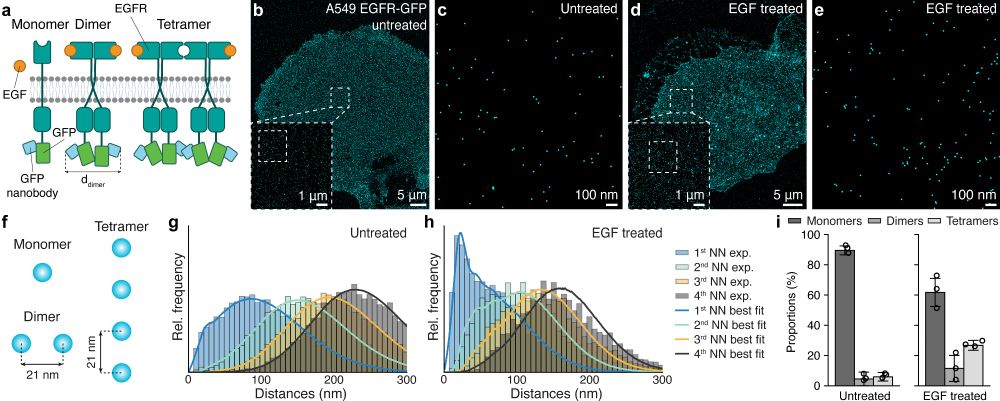
May 7, 2025 at 2:43 PM
We further quantitatively evaluate the oligomerization of the Epidermal Growth Factor Receptor (EGFR) upon binding of its ligand, EGF. 5/7


