Julia Belk
@juliabelk.bsky.social
HHMI Hanna Gray Fellow @StanfordMed
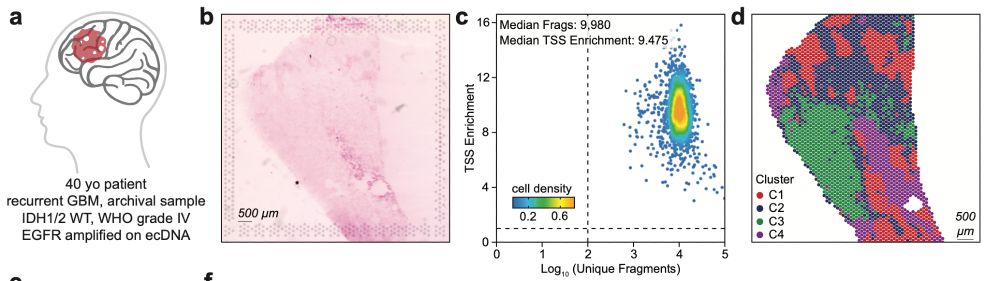
We also apply SPACE-seq to an archived glioblastoma sample, showing that SPACE-seq works well with clinical samples and can detect extrachromosomal DNA amplifications and somatic mutations. (4/5)
April 18, 2025 at 6:36 PM
We also apply SPACE-seq to an archived glioblastoma sample, showing that SPACE-seq works well with clinical samples and can detect extrachromosomal DNA amplifications and somatic mutations. (4/5)
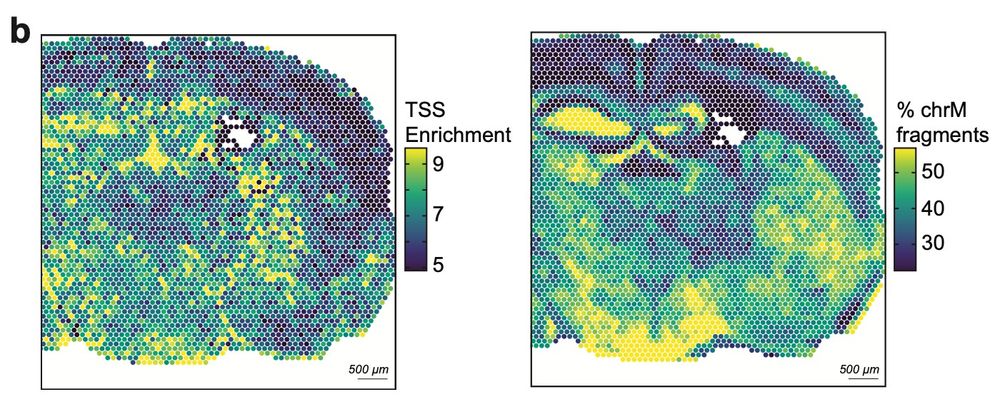
Mitochondrial DNA is captured as well, enabling mtscATAC (a lineage tracing technique previously developed by @caleblareau.bsky.social, @leifludwig.bsky.social, and @bloodgenes.bsky.social) to be applied spatially. (3/5)
April 18, 2025 at 6:36 PM
Mitochondrial DNA is captured as well, enabling mtscATAC (a lineage tracing technique previously developed by @caleblareau.bsky.social, @leifludwig.bsky.social, and @bloodgenes.bsky.social) to be applied spatially. (3/5)
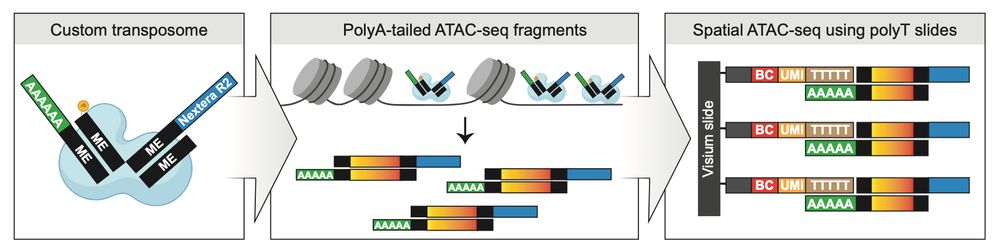
The main idea here is that by tagmenting tissue sections with polyA-tailed adapters, instead of the standard ATAC-seq adapters, we can use spatial transcriptomics slides from @10xgenomics.bsky.social to capture both ATAC and RNA data with spatial resolution. (2/5)
April 18, 2025 at 6:36 PM
The main idea here is that by tagmenting tissue sections with polyA-tailed adapters, instead of the standard ATAC-seq adapters, we can use spatial transcriptomics slides from @10xgenomics.bsky.social to capture both ATAC and RNA data with spatial resolution. (2/5)

