
HudsonAlpha Genome Sequencing Center and DOE Joint Genome Institute
This is something you find when you dig deep enough. We've been looking for ways to harmonize annotations since we saw a similar pattern among pecan genomes in 2021 (buried in the SI tho). www.nature.com/articles/s41...

This is something you find when you dig deep enough. We've been looking for ways to harmonize annotations since we saw a similar pattern among pecan genomes in 2021 (buried in the SI tho). www.nature.com/articles/s41...
IGC-reannotation is not perfect, but reduces this to 5-15%

IGC-reannotation is not perfect, but reduces this to 5-15%
Within two groups that annotated 7 and 23 soybean genomes there were 3x & 2x more PAVs than IGC — these pangenomes are not as 'open' as reported.

Within two groups that annotated 7 and 23 soybean genomes there were 3x & 2x more PAVs than IGC — these pangenomes are not as 'open' as reported.



While PAV generally scales with divergence time, it is 2-4X more common in plants, especially those with a history of polyploids.

While PAV generally scales with divergence time, it is 2-4X more common in plants, especially those with a history of polyploids.
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...
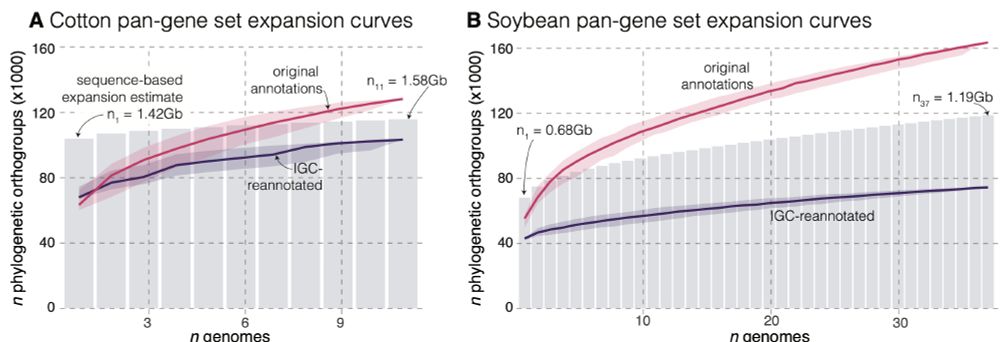
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...





At a continent-wide scale, gene flow was higher between populations in drought-prone habitats ...

At a continent-wide scale, gene flow was higher between populations in drought-prone habitats ...



Here we built 33 reference genomes and short-read resequenced ~1900 varieties that span its genetic diversity.

Here we built 33 reference genomes and short-read resequenced ~1900 varieties that span its genetic diversity.












