Josh Levy
@josh-levy.bsky.social
Virus spread and evolution + connection to behavior.
@Scripps Research, CA.
@Scripps Research, CA.
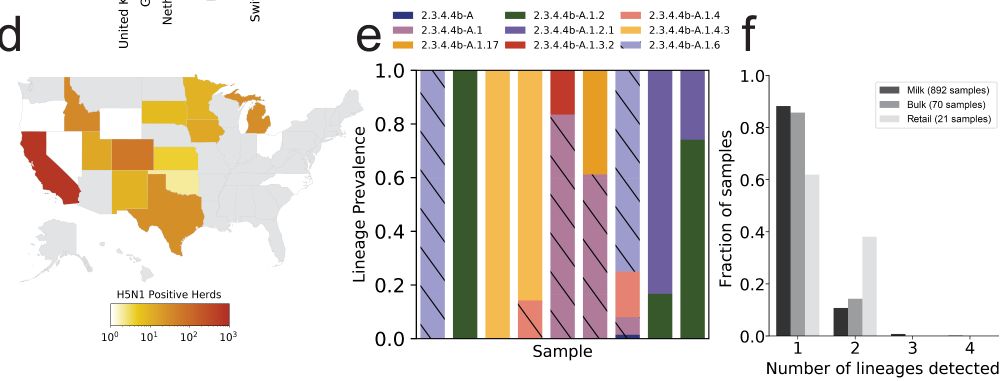
For H5N1, we used publicly available milk sequencing, contributed by USDA and academic labs across the US, derived from “milk” from individual cows, to “bulk tanks” with milk from many cows, up to “retail milk” with milk from many cows and many farms.
July 31, 2025 at 7:18 PM
For H5N1, we used publicly available milk sequencing, contributed by USDA and academic labs across the US, derived from “milk” from individual cows, to “bulk tanks” with milk from many cows, up to “retail milk” with milk from many cows and many farms.
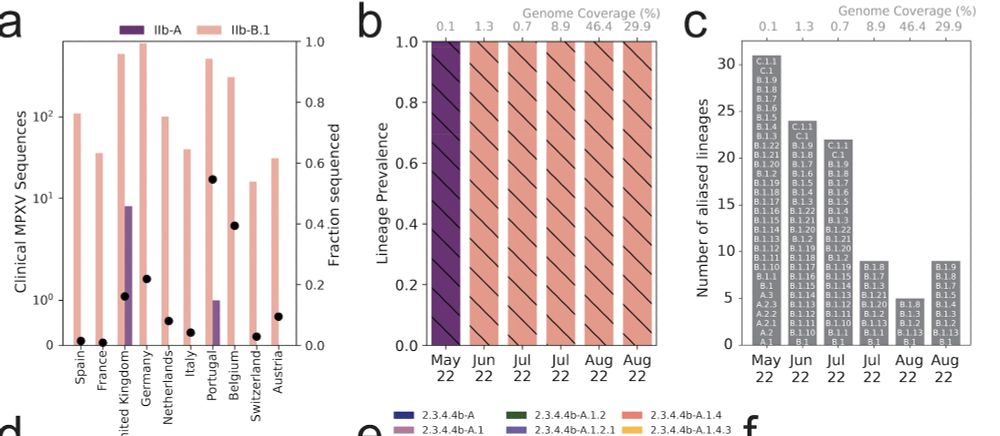
We deployed this approach to track MPXV in Germany and Influenza A/H5N1 sequencing in the US from 2022 and 2024. While we found MPXV sequencing didn't recover lineage mixtures, Freyja 2 uncovered candidate sublineages (not resolvable due to low genomic coverage) that were likely circulating.
July 31, 2025 at 7:18 PM
We deployed this approach to track MPXV in Germany and Influenza A/H5N1 sequencing in the US from 2022 and 2024. While we found MPXV sequencing didn't recover lineage mixtures, Freyja 2 uncovered candidate sublineages (not resolvable due to low genomic coverage) that were likely circulating.
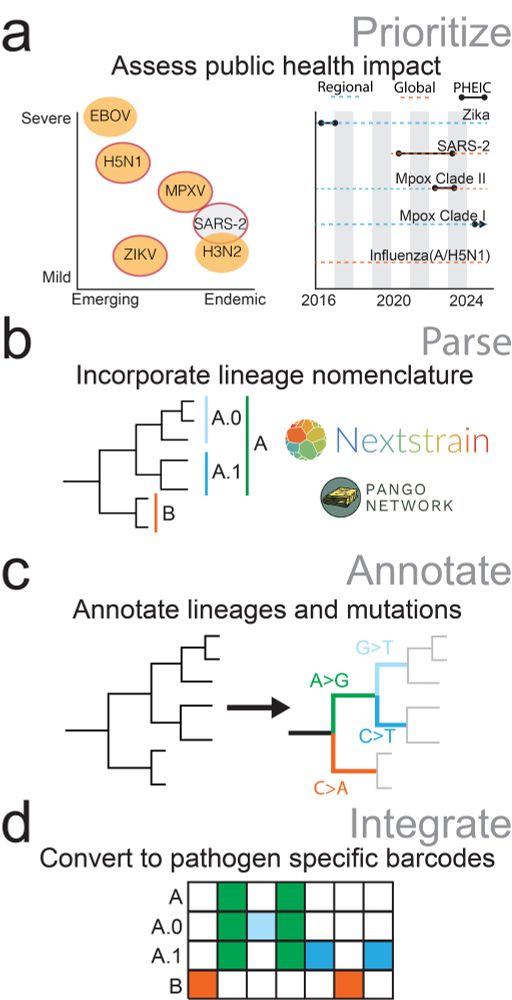
Our next step was to generalize this approach to other pathogens. To do this, we built workflows to extract lineage-defining mutation definitions, and convert those into “barcode” format for Freyja use. We’ve done this for MPXV, RSV, H5N1, and more, and our workflow is freely available.
July 31, 2025 at 7:18 PM
Our next step was to generalize this approach to other pathogens. To do this, we built workflows to extract lineage-defining mutation definitions, and convert those into “barcode” format for Freyja use. We’ve done this for MPXV, RSV, H5N1, and more, and our workflow is freely available.
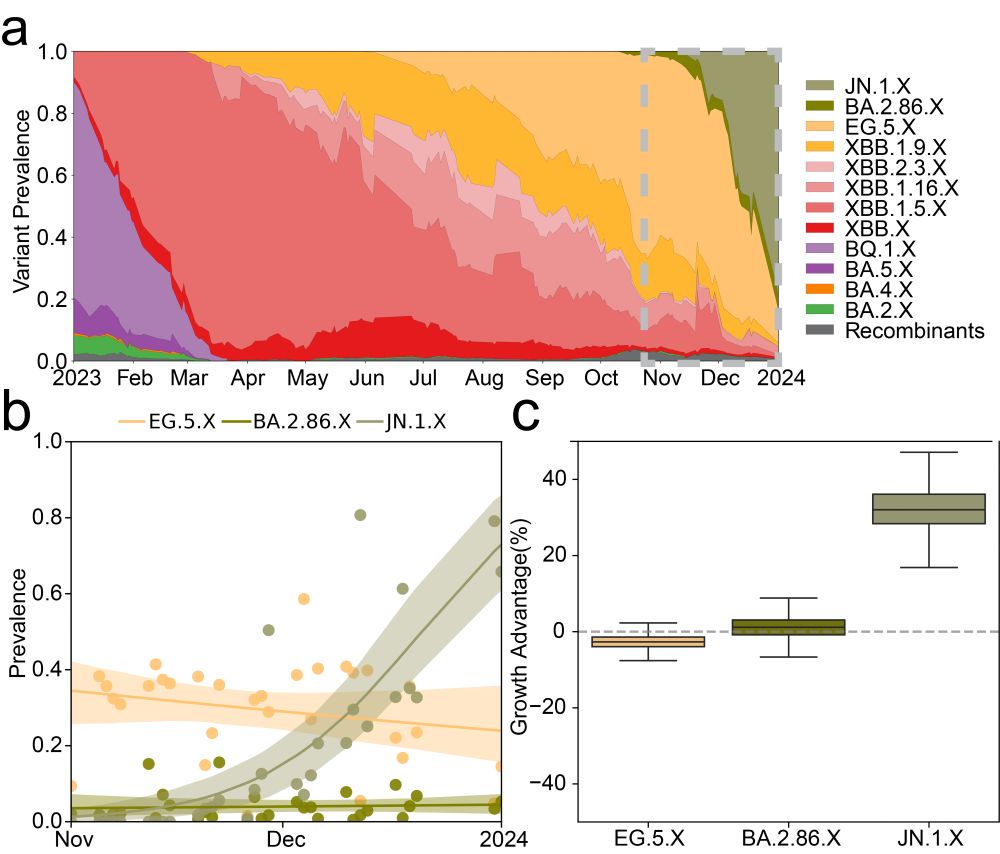
We also incorporated a method to calculate lineage-specific growth advantage from longitudinal wastewater sequencing. Using data collected as part of SEARCH efforts in San Diego, we found Freyja 2 recovers growth advantages including for JN.1 that match literature results from clinical estimates.
July 31, 2025 at 7:18 PM
We also incorporated a method to calculate lineage-specific growth advantage from longitudinal wastewater sequencing. Using data collected as part of SEARCH efforts in San Diego, we found Freyja 2 recovers growth advantages including for JN.1 that match literature results from clinical estimates.
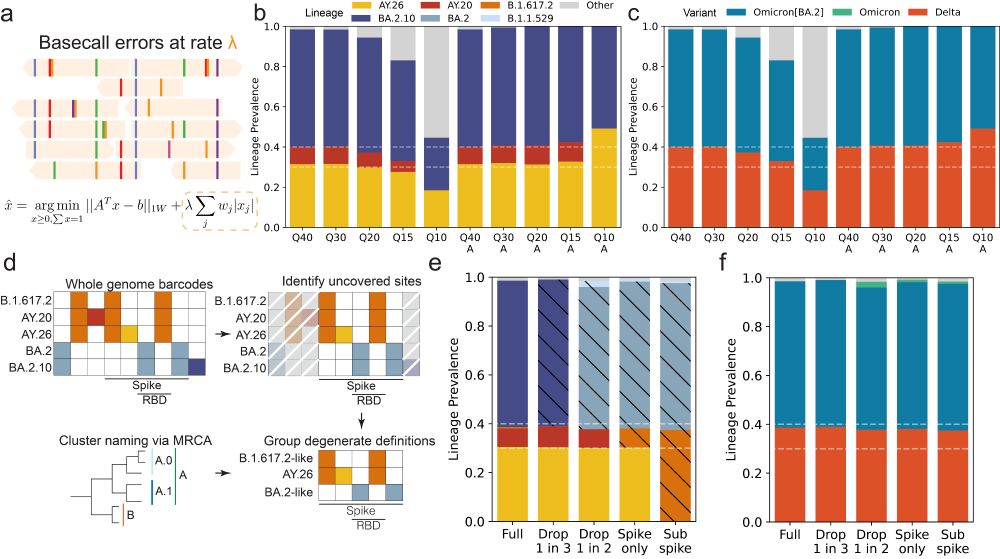
Noting the many approaches and platforms used to sequence SARS-CoV-2 in wastewater, we established functionality that accounts for incomplete coverage in sequencing (making some lineages indistinguishable), and adaptive regularization that allows for Freyja 2 to account for empirical error rates.
July 31, 2025 at 7:18 PM
Noting the many approaches and platforms used to sequence SARS-CoV-2 in wastewater, we established functionality that accounts for incomplete coverage in sequencing (making some lineages indistinguishable), and adaptive regularization that allows for Freyja 2 to account for empirical error rates.
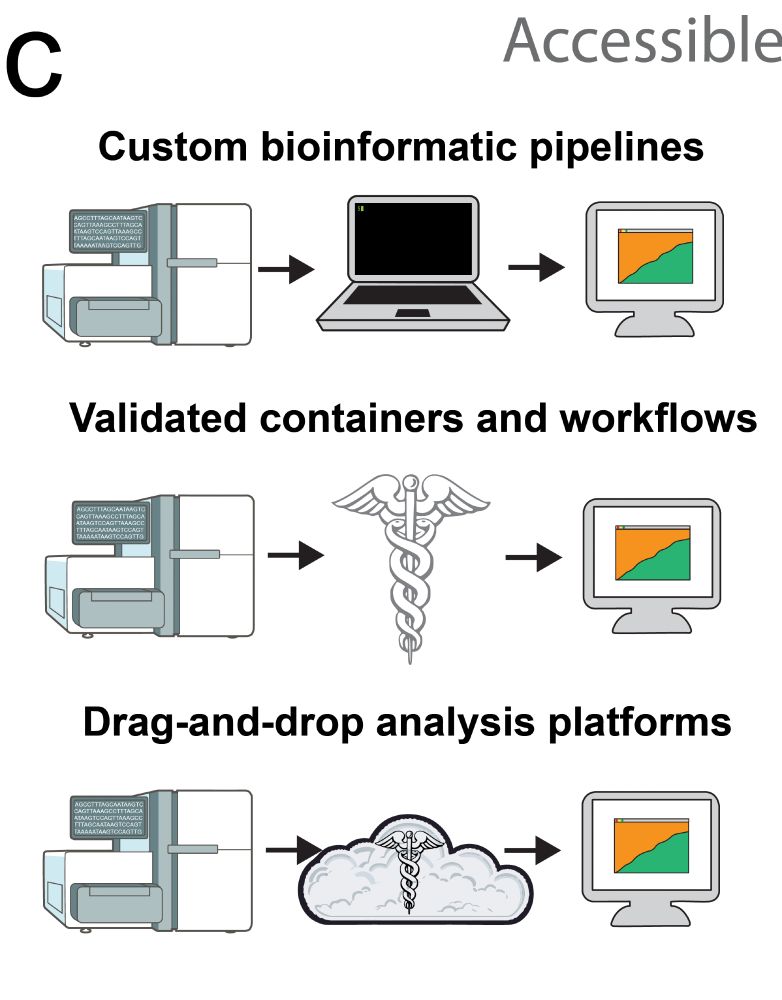
Working with awesome groups like Theiagen and STaPH-B, we made Freyja 2 accessible to a broad range of users, including through drag and drop automated result generation via Terra/Galaxy (great for labs with limited bioinformatics experience), conda, and Docker containers.
July 31, 2025 at 7:18 PM
Working with awesome groups like Theiagen and STaPH-B, we made Freyja 2 accessible to a broad range of users, including through drag and drop automated result generation via Terra/Galaxy (great for labs with limited bioinformatics experience), conda, and Docker containers.
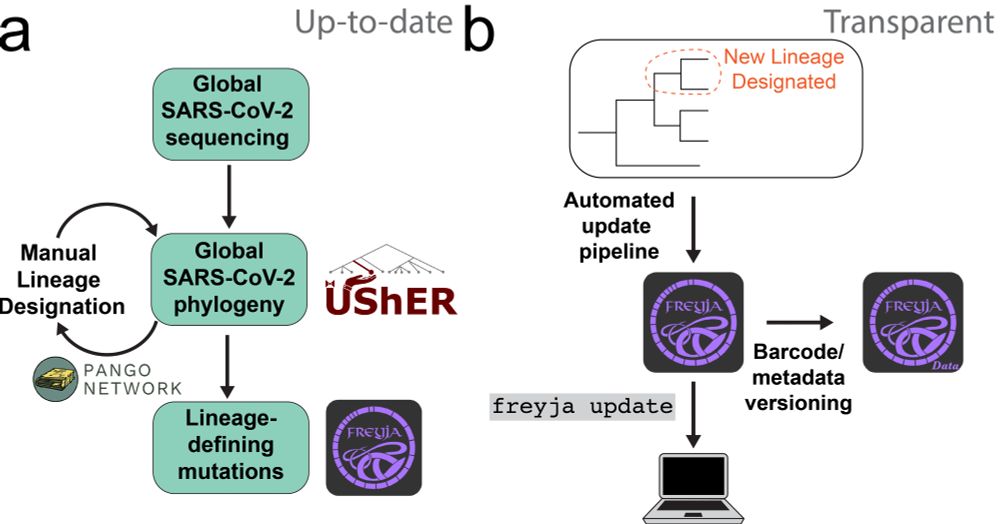
Now to the paper: By leveraging the wealth of amazing resources for tracking the evolution of SARS-CoV-2, we established automated pipelines that keep analyses in line with the most recent pango designations, and allow users to access that information effectively immediately.
July 31, 2025 at 7:18 PM
Now to the paper: By leveraging the wealth of amazing resources for tracking the evolution of SARS-CoV-2, we established automated pipelines that keep analyses in line with the most recent pango designations, and allow users to access that information effectively immediately.
Standing up for science over in here in San Diego #standupforscience2025

March 7, 2025 at 9:53 PM
Standing up for science over in here in San Diego #standupforscience2025

