
Jon Mifsud
@jonathonmifsud.bsky.social
Postdoc @ KU Leuven | Virus evolution 🧬👨💻
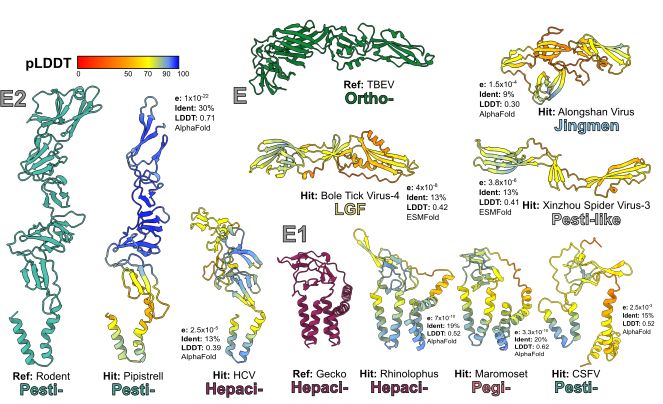
Lastly, we then dive further into a group of LGFs, the Bole Tick Viruses, which might represent emerging threat to human health and examine the distribution of other proteins and enzymes (e.g., Erns, MTase and prM) across the Flaviviridae.

February 7, 2024 at 11:02 AM
Lastly, we then dive further into a group of LGFs, the Bole Tick Viruses, which might represent emerging threat to human health and examine the distribution of other proteins and enzymes (e.g., Erns, MTase and prM) across the Flaviviridae.
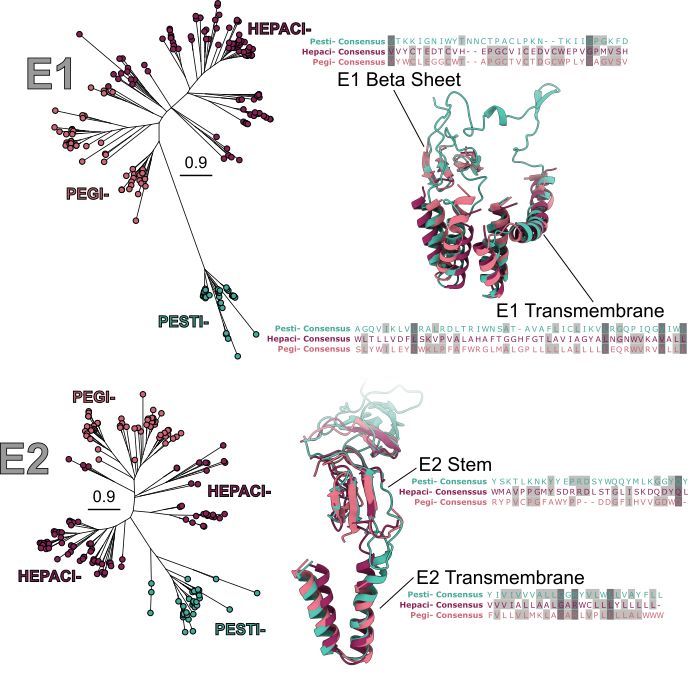
Conversely, Lineage 2 shows less diversity, with Hepaci-/Pegiviruses having similar genome organisation and greater conservation in RdRp and E1E2 sequences and structures.
February 7, 2024 at 10:59 AM
Conversely, Lineage 2 shows less diversity, with Hepaci-/Pegiviruses having similar genome organisation and greater conservation in RdRp and E1E2 sequences and structures.
Lineage 1 of Flaviviridae diversified extensively: Jingmenviruses segmented and lost a fusion loop; Orthoflaviviruses gained prM; LGF and Pestiviruses got RNase T2, likely from bacteria.

February 7, 2024 at 10:58 AM
Lineage 1 of Flaviviridae diversified extensively: Jingmenviruses segmented and lost a fusion loop; Orthoflaviviruses gained prM; LGF and Pestiviruses got RNase T2, likely from bacteria.
Mapping glycoprotein structures across Flaviviridae (+structural 🌳s) identifies two lineages: The 1st includes Orthoflavivirus/Jingmenvirus and LGF/Pestivirus clades, originating from an ancestor with an E glycoprotein and cap-dependent translation, requiring a methyltransferase

February 7, 2024 at 10:56 AM
Mapping glycoprotein structures across Flaviviridae (+structural 🌳s) identifies two lineages: The 1st includes Orthoflavivirus/Jingmenvirus and LGF/Pestivirus clades, originating from an ancestor with an E glycoprotein and cap-dependent translation, requiring a methyltransferase
The breadth of the Flaviviridae has grown enormously with metagenomic studies and with it what we know as a “flavivirus”. So how did these two glycoprotein systems map across the enormous diversity of flaviviruses, and how might these systems have arisen?
February 7, 2024 at 10:56 AM
The breadth of the Flaviviridae has grown enormously with metagenomic studies and with it what we know as a “flavivirus”. So how did these two glycoprotein systems map across the enormous diversity of flaviviruses, and how might these systems have arisen?
We then folded 621 polyprotein sequences with AlphaFold & ESMFold. AlphaFold excelled for most, but where MSA depth decreased (e.g., the poorly sampled LGF) we found that ESMFold, which doesn’t require MSA performed better for some taxa! (more on this in the 📄)

February 7, 2024 at 10:54 AM
We then folded 621 polyprotein sequences with AlphaFold & ESMFold. AlphaFold excelled for most, but where MSA depth decreased (e.g., the poorly sampled LGF) we found that ESMFold, which doesn’t require MSA performed better for some taxa! (more on this in the 📄)
We started off collating and curating all Flaviviridae genomes, then built RdRp trees (225 in total), forming our study's backbone. We find that the Flaviviridae can be broken up into three clades: 1) LGF & Pestivirus, 2) Orthoflavivirus-Jingmenvirus, and 3) Pegivirus-Hepacivirus

February 7, 2024 at 10:54 AM
We started off collating and curating all Flaviviridae genomes, then built RdRp trees (225 in total), forming our study's backbone. We find that the Flaviviridae can be broken up into three clades: 1) LGF & Pestivirus, 2) Orthoflavivirus-Jingmenvirus, and 3) Pegivirus-Hepacivirus
Glycoproteins are crucial for virus function, yet, due to homology and alignment challenges, we often focus on conserved proteins like RdRp. We can now overcome these problems with protein structure prediction, shedding light on Flaviviridae's evolution!

February 7, 2024 at 10:52 AM
Glycoproteins are crucial for virus function, yet, due to homology and alignment challenges, we often focus on conserved proteins like RdRp. We can now overcome these problems with protein structure prediction, shedding light on Flaviviridae's evolution!
New paper alert🚨 We've mapped glycoproteins across Flaviviridae using phylogenetics & machine-learning-enabled protein structure prediction, uncovering complex evolutionary history shaping this family's virology & ecology. 🧵 ⬇️
Preprint: www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...

February 7, 2024 at 10:51 AM
New paper alert🚨 We've mapped glycoproteins across Flaviviridae using phylogenetics & machine-learning-enabled protein structure prediction, uncovering complex evolutionary history shaping this family's virology & ecology. 🧵 ⬇️
Preprint: www.biorxiv.org/content/10.1...
Preprint: www.biorxiv.org/content/10.1...

