
We provide suggestions to "mitigate the reporting of false discoveries that, if left unchecked, would undermine confidence in large-scale perturbational screens."

We provide suggestions to "mitigate the reporting of false discoveries that, if left unchecked, would undermine confidence in large-scale perturbational screens."




We found the best performance if we considered only 1 mismatch in the SDR, at a CFD sum of 4.8, so we define that as our "CRISPick Aggregate CFD" threshold

We found the best performance if we considered only 1 mismatch in the SDR, at a CFD sum of 4.8, so we define that as our "CRISPick Aggregate CFD" threshold




Good to know a search engine thinks a jar of mayonnaise is a better fit for mayor of my city than some nepo baby.
Good to know a search engine thinks a jar of mayonnaise is a better fit for mayor of my city than some nepo baby.





screen should continue to be a standard practice to reduce false positives and to increase confidence in true hits..."

screen should continue to be a standard practice to reduce false positives and to increase confidence in true hits..."

esahubble.org/images/potw2...

esahubble.org/images/potw2...



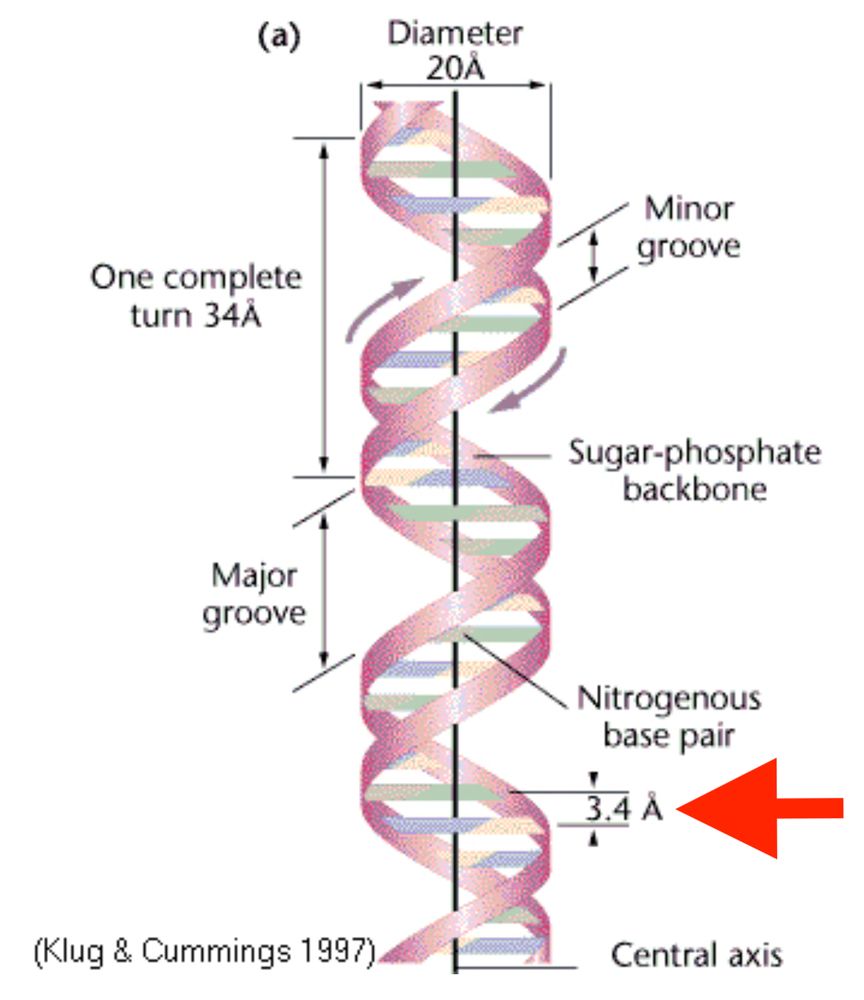

And no, I did not actually hit send, as much as I wanted to.

And no, I did not actually hit send, as much as I wanted to.

