MADRe is open-source, modular, and ready to use.
Check it out:
🔗 github.com/lbcb-sci/MADRe
9/9

MADRe is open-source, modular, and ready to use.
Check it out:
🔗 github.com/lbcb-sci/MADRe
9/9
While low-abundance strains may be underrepresented, this trade-off significantly reduces false-positive identifications, a common issue in strain-level metagenomics.
8/9
While low-abundance strains may be underrepresented, this trade-off significantly reduces false-positive identifications, a common issue in strain-level metagenomics.
8/9
✅ Comparable or improved accuracy over existing tools
✅ Clearer and more realistic abundance profiles
✅ Substantial reductions in runtime and memory usage
7/9

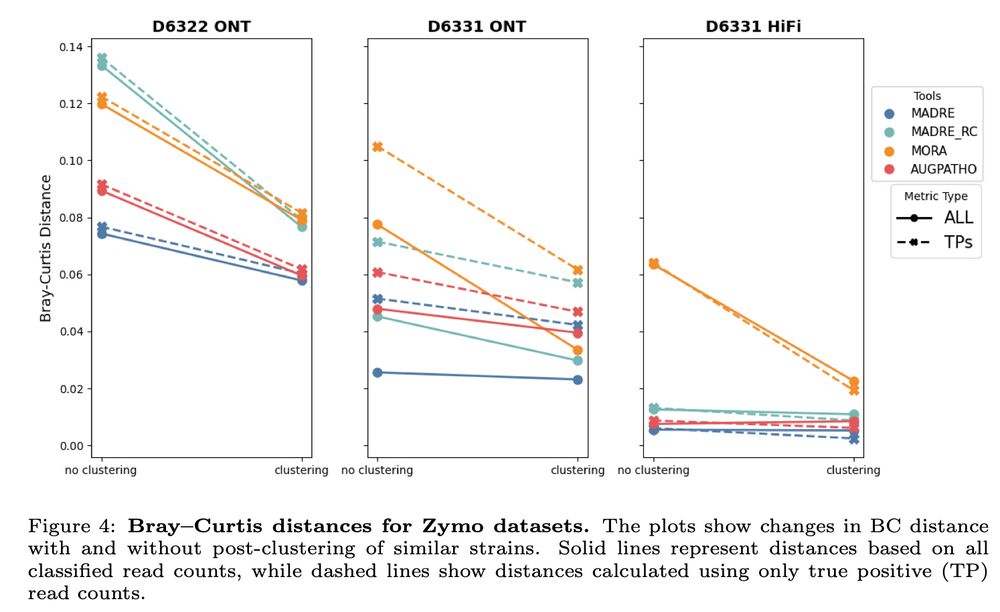
✅ Comparable or improved accuracy over existing tools
✅ Clearer and more realistic abundance profiles
✅ Substantial reductions in runtime and memory usage
7/9

This enhances classification sensitivity and filters false-positive identifications, enabling precise strain-level profiling. 5/9

This enhances classification sensitivity and filters false-positive identifications, enabling precise strain-level profiling. 5/9
However, this step alone does not eliminate all false positives and does not provide accurate abundance estimates. 4/9
However, this step alone does not eliminate all false positives and does not provide accurate abundance estimates. 4/9
Using EM-based read reassignment and info about strain collapses, we construct reduced database. 3/9
Using EM-based read reassignment and info about strain collapses, we construct reduced database. 3/9
However, mapping reads to such large databases is computationally expensive and often impractical at scale. 2/9
However, mapping reads to such large databases is computationally expensive and often impractical at scale. 2/9

