
James Lloyd 🧬
@jamespblloyd.bsky.social
Synthetic AstroBotatnist. Engineer of synthetic gene circuits in plants. 🇬🇧🇪🇺 now in 🇦🇺. He/They.
Google Scholar: http://shorturl.at/dnHVZ
Compbio blog: badgrammargoodsyntax.com
Google Scholar: http://shorturl.at/dnHVZ
Compbio blog: badgrammargoodsyntax.com
I’ve only had time to review grants, no papers, this year because I refuse to outsource it to AI and I have been overwhelmed.
Not proud to be slacking my duties. But grants reviewing is a requirement for me, papers are optional. I hope to return to the duty next year.
Not proud to be slacking my duties. But grants reviewing is a requirement for me, papers are optional. I hope to return to the duty next year.
November 11, 2025 at 11:26 AM
I’ve only had time to review grants, no papers, this year because I refuse to outsource it to AI and I have been overwhelmed.
Not proud to be slacking my duties. But grants reviewing is a requirement for me, papers are optional. I hope to return to the duty next year.
Not proud to be slacking my duties. But grants reviewing is a requirement for me, papers are optional. I hope to return to the duty next year.
This computational work was possible because of the great team of Moji, Pratosh & Georgina. Also building on the great computational tool TranSuite (pre-print below):
www.biorxiv.org/content/10.1...
This is a big problem in translation prediction, picking the wrong ORF, but is too often overlooked
www.biorxiv.org/content/10.1...
This is a big problem in translation prediction, picking the wrong ORF, but is too often overlooked
November 6, 2025 at 2:14 PM
This computational work was possible because of the great team of Moji, Pratosh & Georgina. Also building on the great computational tool TranSuite (pre-print below):
www.biorxiv.org/content/10.1...
This is a big problem in translation prediction, picking the wrong ORF, but is too often overlooked
www.biorxiv.org/content/10.1...
This is a big problem in translation prediction, picking the wrong ORF, but is too often overlooked
Because automated protein structure prediction is becoming more common, I wondered what impact this would have on protein structure predictions. Taking the "wrong" ORF indicated a longer predicted protein, missing a small part of the C-terminal. But the reality is just a small protein would be made!

November 6, 2025 at 2:14 PM
Because automated protein structure prediction is becoming more common, I wondered what impact this would have on protein structure predictions. Taking the "wrong" ORF indicated a longer predicted protein, missing a small part of the C-terminal. But the reality is just a small protein would be made!
To show that this improvement extended beyond just two example genes I noticed manually, we re-examined some old RNA-seq from an NMD mutant in Arabidopsis. Sure enough, known NMD features were higher in the revised start/stop (open reading frame; ORF) annotations.

November 6, 2025 at 2:14 PM
To show that this improvement extended beyond just two example genes I noticed manually, we re-examined some old RNA-seq from an NMD mutant in Arabidopsis. Sure enough, known NMD features were higher in the revised start/stop (open reading frame; ORF) annotations.
So it started when I was showing a visiting High School student the Arabidopsis genome and noticed an error. The wrong start and stop codon was labelled for some NMD targets (PTB1 & RS2Z33).
Using TranSuite (modified & updated) we showed that the biologically correct start/stop were now annotated!
Using TranSuite (modified & updated) we showed that the biologically correct start/stop were now annotated!

November 6, 2025 at 2:14 PM
So it started when I was showing a visiting High School student the Arabidopsis genome and noticed an error. The wrong start and stop codon was labelled for some NMD targets (PTB1 & RS2Z33).
Using TranSuite (modified & updated) we showed that the biologically correct start/stop were now annotated!
Using TranSuite (modified & updated) we showed that the biologically correct start/stop were now annotated!
This is a fantastic extension of the memory circuits for plants, and shows how much hard work is needed to de-bug unexpected problems when faced in synthetic biology. This is a testament to the determination and creative of Patrick and the team. I am beyond proud to work with such fantastic people.
October 31, 2025 at 9:24 AM
This is a fantastic extension of the memory circuits for plants, and shows how much hard work is needed to de-bug unexpected problems when faced in synthetic biology. This is a testament to the determination and creative of Patrick and the team. I am beyond proud to work with such fantastic people.
Patrick also showed split-recombinases can work with new bioconjugation domains. Then he went big and showed that a 6-input AND gate with a >100 fold induction was possible by using three split recombinases in the same circuit, pushing the limits of computation in plant cells!
October 31, 2025 at 9:24 AM
Patrick also showed split-recombinases can work with new bioconjugation domains. Then he went big and showed that a 6-input AND gate with a >100 fold induction was possible by using three split recombinases in the same circuit, pushing the limits of computation in plant cells!
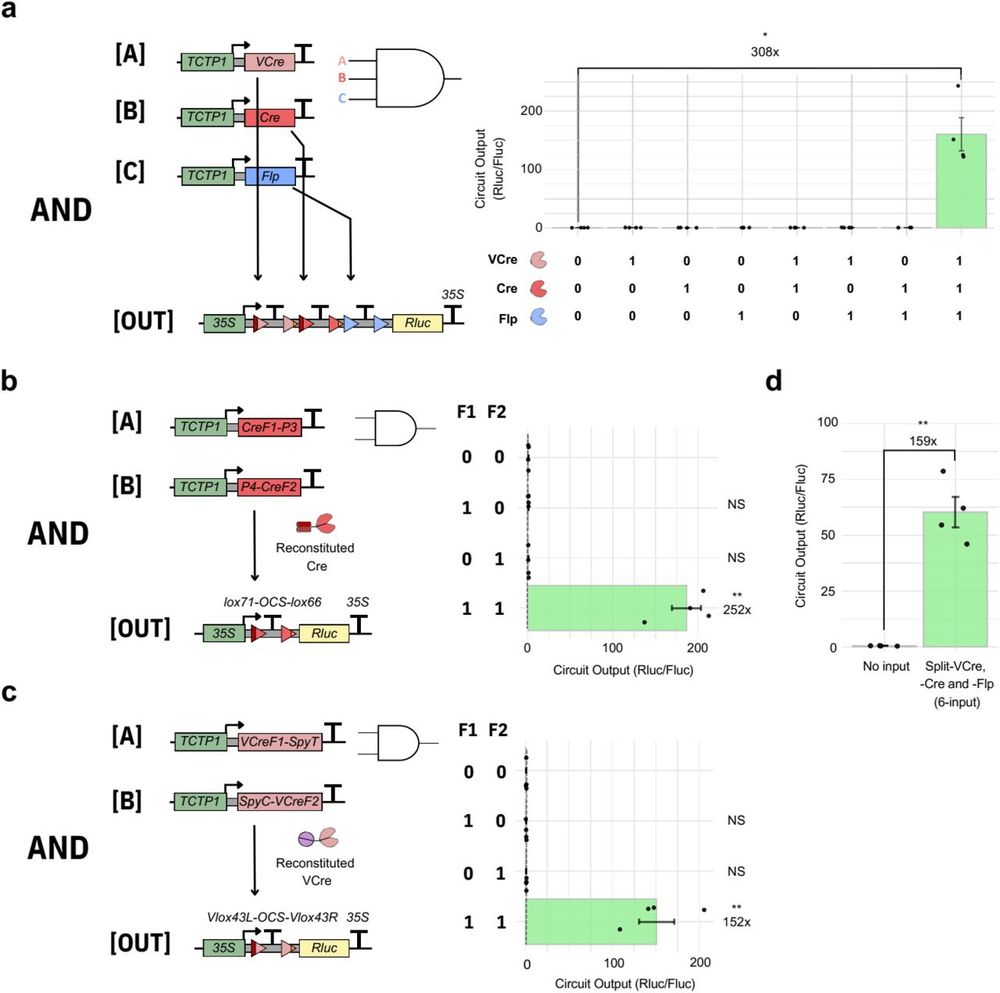
Patrick, not satisfied with the 2-input AND gate we had published in 2022, designed & built a 3-input AND gate with the new recombinases he had de-bugged, seeing a >300 activation in the 3-input on-state and negligible noise in all other conditions. Fantastic.
October 31, 2025 at 9:24 AM
Patrick, not satisfied with the 2-input AND gate we had published in 2022, designed & built a 3-input AND gate with the new recombinases he had de-bugged, seeing a >300 activation in the 3-input on-state and negligible noise in all other conditions. Fantastic.
Additionally, circuit performance was tested not only in Arabidopsis, the model plant, but also in tomato! So we can see these circuits can function in a crop as well!
October 31, 2025 at 9:24 AM
Additionally, circuit performance was tested not only in Arabidopsis, the model plant, but also in tomato! So we can see these circuits can function in a crop as well!
And with all we had learned, Patrick expanded our toolkit of recombinases, bringing VCre & SCre back into the roster, after I had rejected them in the 2022 study. This builds a strong foundation to allow us to build new, more complex circuits.

October 31, 2025 at 9:24 AM
And with all we had learned, Patrick expanded our toolkit of recombinases, bringing VCre & SCre back into the roster, after I had rejected them in the 2022 study. This builds a strong foundation to allow us to build new, more complex circuits.
Importantly, similar genetic parts for circuits cannot be assumed to behave the same!
Unexpected differences can have disastrous effects when constructing a circuit. Deep characterisation of parts, as done here, is needed to better understand which is best to use in which context.
Unexpected differences can have disastrous effects when constructing a circuit. Deep characterisation of parts, as done here, is needed to better understand which is best to use in which context.
October 31, 2025 at 9:24 AM
Importantly, similar genetic parts for circuits cannot be assumed to behave the same!
Unexpected differences can have disastrous effects when constructing a circuit. Deep characterisation of parts, as done here, is needed to better understand which is best to use in which context.
Unexpected differences can have disastrous effects when constructing a circuit. Deep characterisation of parts, as done here, is needed to better understand which is best to use in which context.
This goes to show that few pairs of Recombinase-Target sites behave the same. FLP site enhances, but the B3 protein enhances. The Cre family all interfere, but only the lox site has an inhibitory effect - I guess we just got epically unlucky with pairing that recombinase with that promoter!

October 31, 2025 at 9:24 AM
This goes to show that few pairs of Recombinase-Target sites behave the same. FLP site enhances, but the B3 protein enhances. The Cre family all interfere, but only the lox site has an inhibitory effect - I guess we just got epically unlucky with pairing that recombinase with that promoter!
Then Patrick tested if Act2 was affected by other recombinase target sites: B3, VCre & SCre sites had no effect.
But FLP target site augmented expression!
What about the protein level? Cre interferes, also SCre & VCre. FLP did nothing. B3 enhances expression. A cryptic transcription factor?
But FLP target site augmented expression!
What about the protein level? Cre interferes, also SCre & VCre. FLP did nothing. B3 enhances expression. A cryptic transcription factor?
October 31, 2025 at 9:24 AM
Then Patrick tested if Act2 was affected by other recombinase target sites: B3, VCre & SCre sites had no effect.
But FLP target site augmented expression!
What about the protein level? Cre interferes, also SCre & VCre. FLP did nothing. B3 enhances expression. A cryptic transcription factor?
But FLP target site augmented expression!
What about the protein level? Cre interferes, also SCre & VCre. FLP did nothing. B3 enhances expression. A cryptic transcription factor?
Patrick then tested lox negation w/ a range of promoters. Act2 was repressed, but so too was TCTP & Ubi10. Only 35S & NOS did not experience negation from a lox site. Indicating interesting PRO+lox site interactions…is this common in non-plants? I think people better start checking ASAP.

October 31, 2025 at 9:24 AM
Patrick then tested lox negation w/ a range of promoters. Act2 was repressed, but so too was TCTP & Ubi10. Only 35S & NOS did not experience negation from a lox site. Indicating interesting PRO+lox site interactions…is this common in non-plants? I think people better start checking ASAP.
In my original study, I shied away from 35S promoter, given fears reviewers would condemn it, but that the OCS terminator might not repress the transcriptional output from 35S, but Patrick found that the off-state with 35S running immediately into the OCS terminator was still very effective.
October 31, 2025 at 9:24 AM
In my original study, I shied away from 35S promoter, given fears reviewers would condemn it, but that the OCS terminator might not repress the transcriptional output from 35S, but Patrick found that the off-state with 35S running immediately into the OCS terminator was still very effective.


