Investigator @Gladstone Institutes and Parker Institute for Cancer Immunotherapy
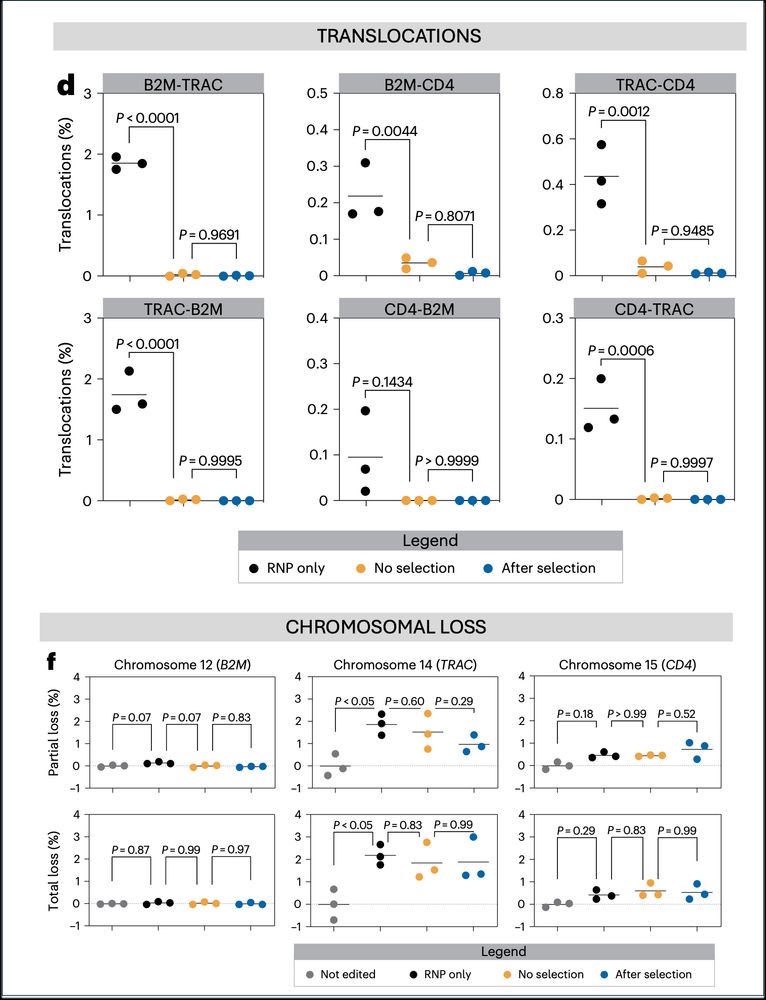
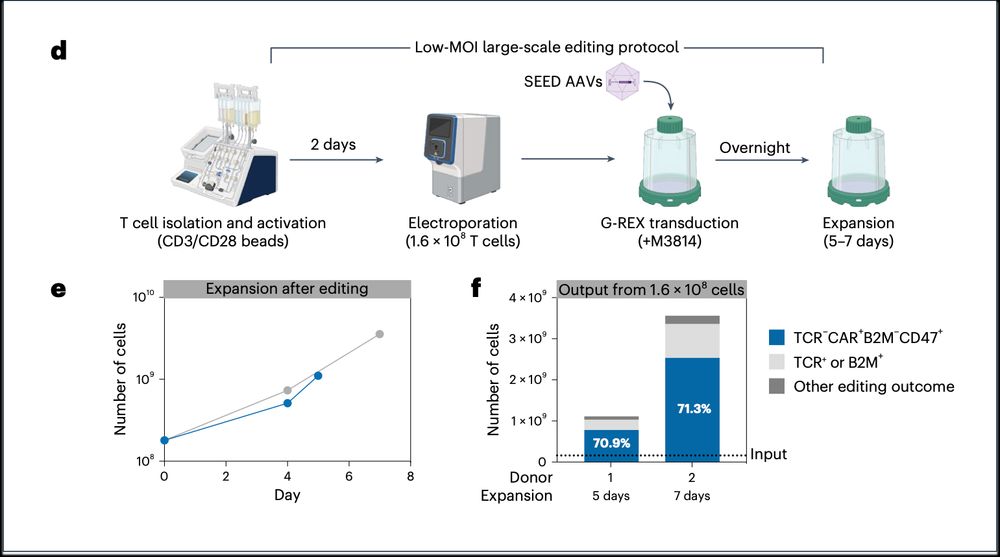
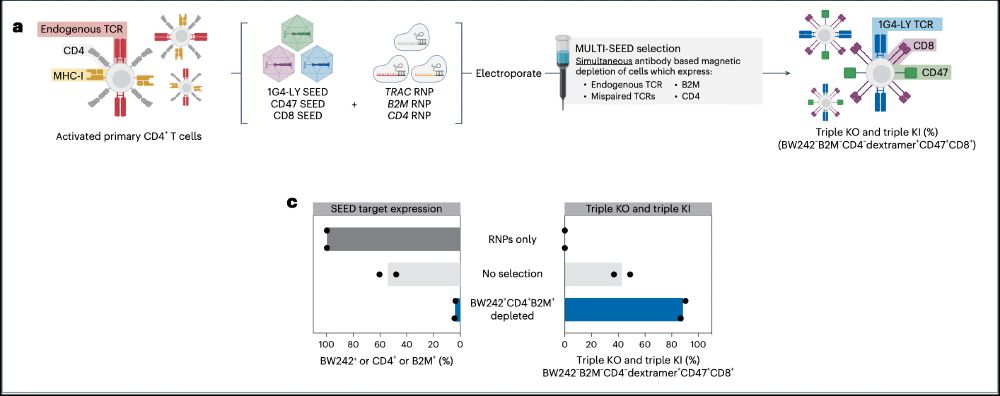
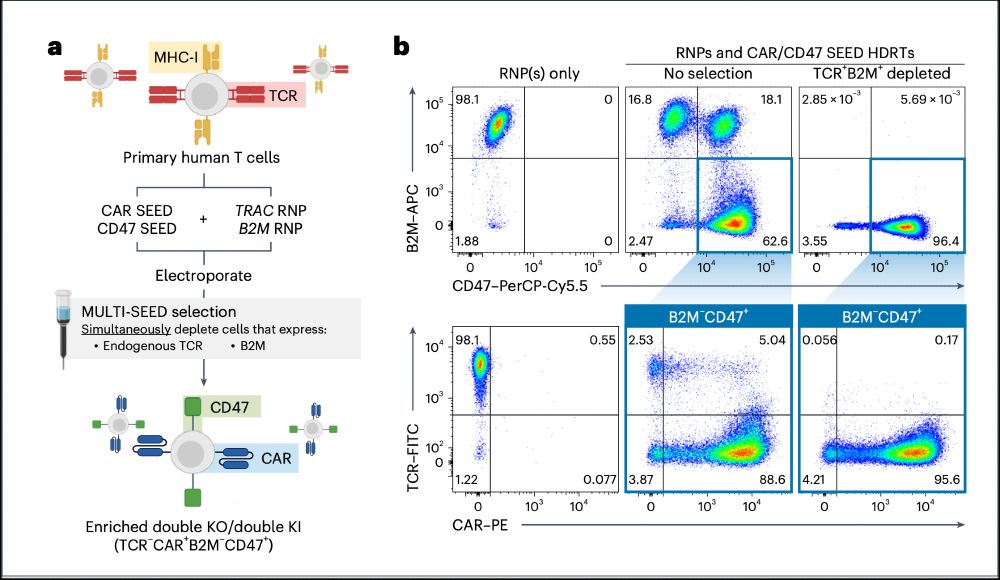
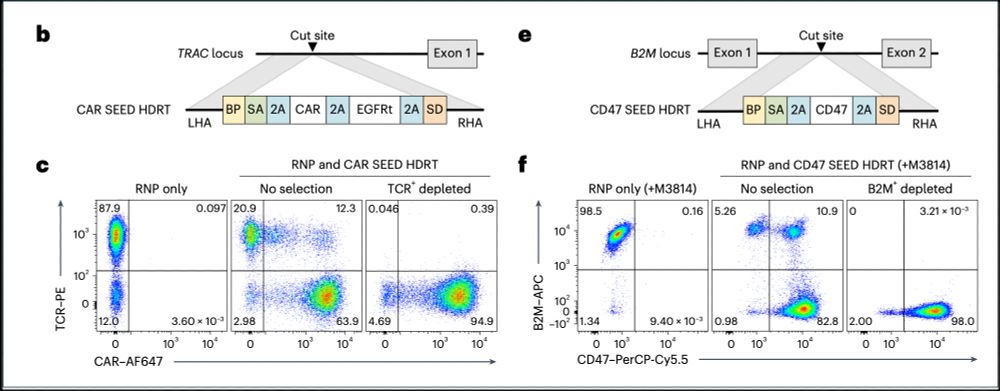
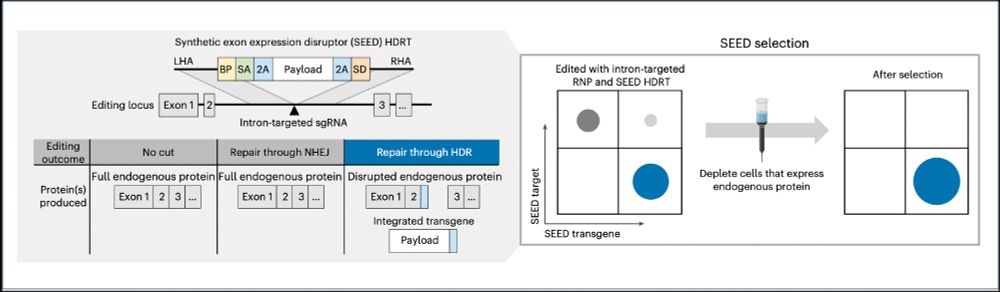
➡️By integrating the epitope-edited TCR, we can identify and eliminate mispaired TCRs and SEED select for cells with proper receptor expression and function!
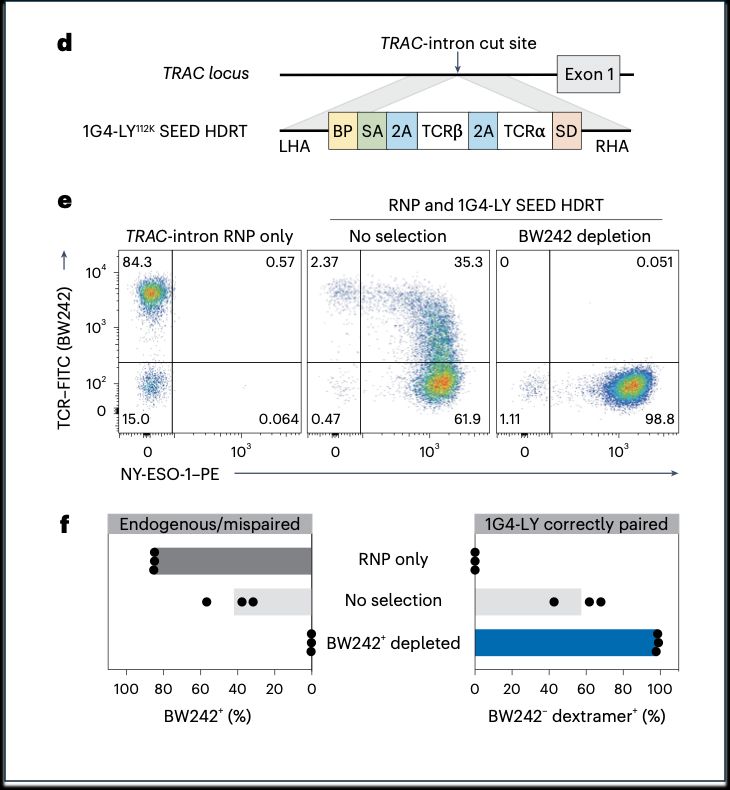
➡️By integrating the epitope-edited TCR, we can identify and eliminate mispaired TCRs and SEED select for cells with proper receptor expression and function!
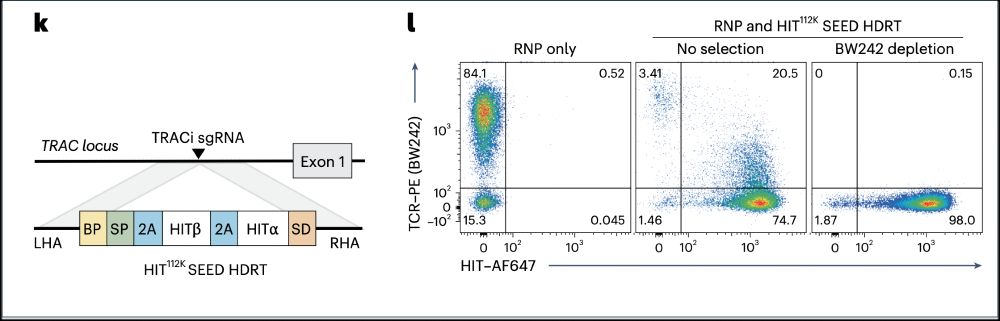
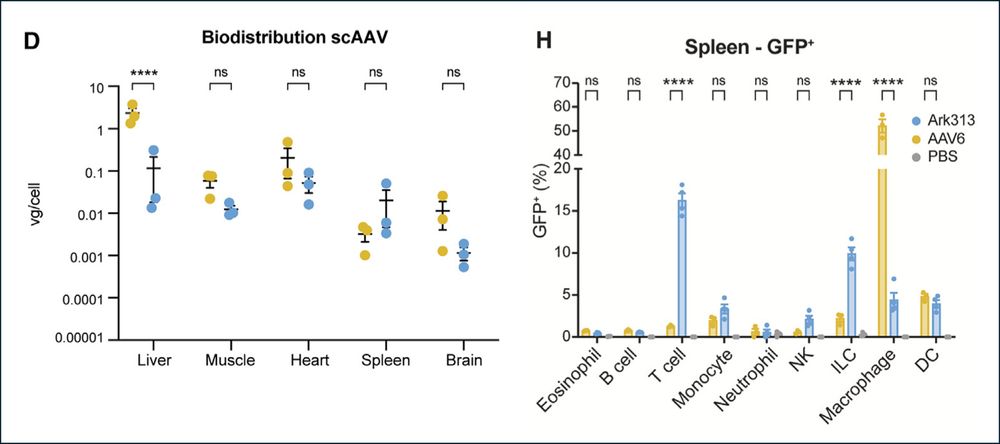
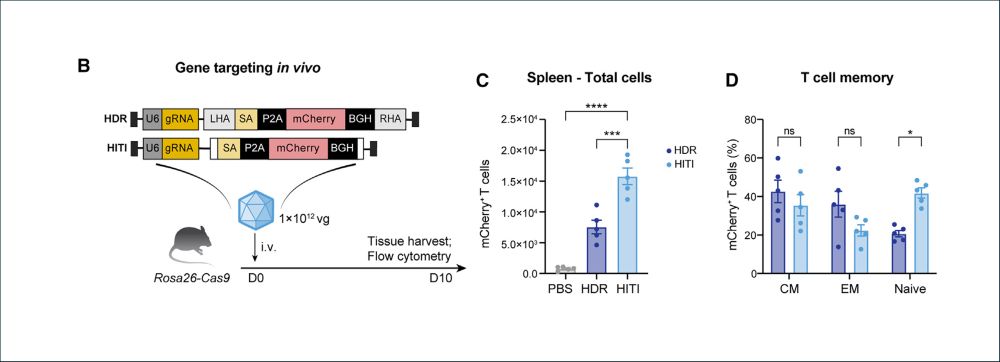
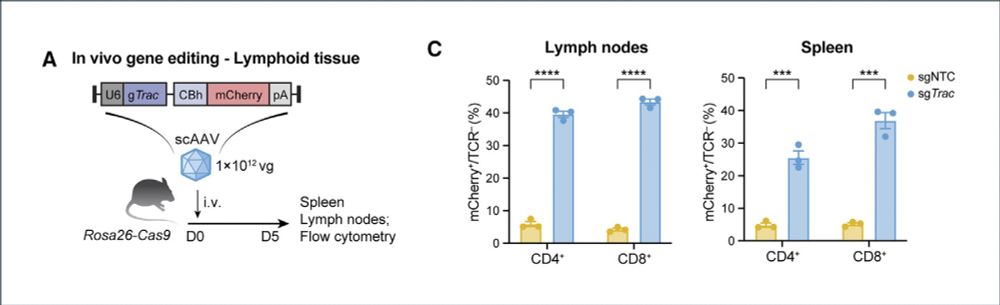
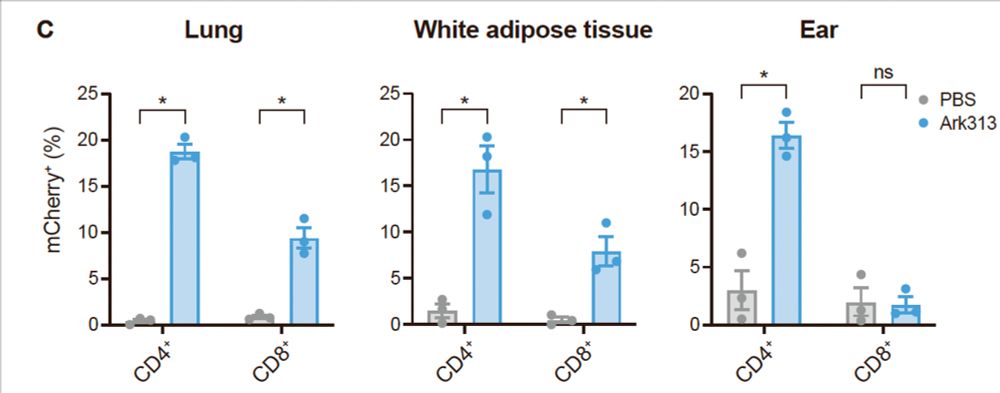
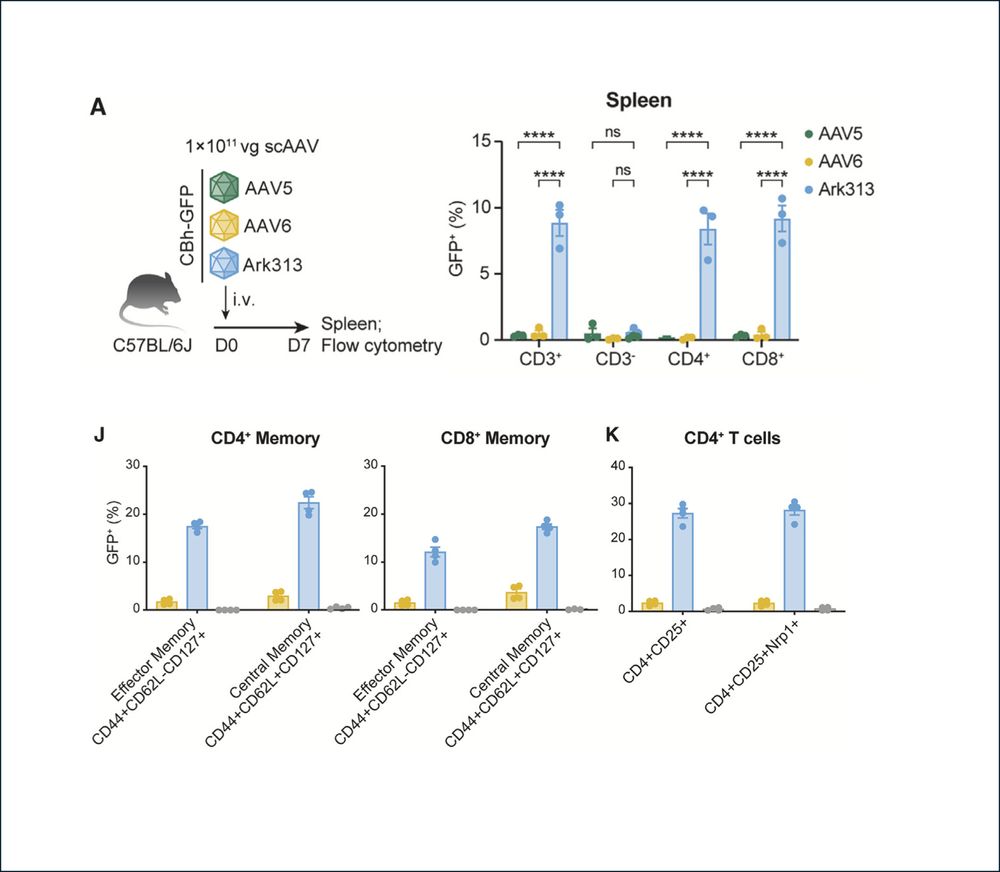
🔥In vivo gene editing to enhance anti-tumor immunity!
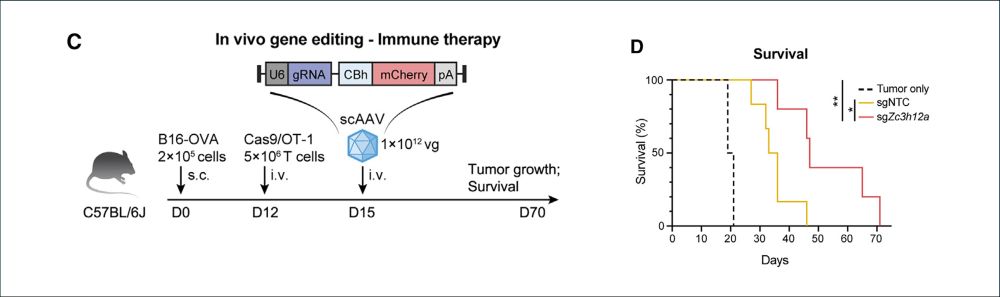
🔥In vivo gene editing to enhance anti-tumor immunity!