
Hi-C in cells depleted of PDS5 & now partially of NIPBL recover some of CTCF boundaries (+ also compartments)

Hi-C in cells depleted of PDS5 & now partially of NIPBL recover some of CTCF boundaries (+ also compartments)
PDS5+NIPBL could colocalize on cohesin leading to NIPBL unbinding! Loops often released but sometimes shrunk & NIPBL returned.
So PDS5 slows loop growth + speeds NIPBL unbinding by facilitated dissociation!

PDS5+NIPBL could colocalize on cohesin leading to NIPBL unbinding! Loops often released but sometimes shrunk & NIPBL returned.
So PDS5 slows loop growth + speeds NIPBL unbinding by facilitated dissociation!
PDS5 inhibits cohesin-NIPBL ATPase+binding in vitro & suppresses NIPBL occupancy of cohesin in cells(+ PDS5&WAPL dont affect each others occupancy)
iFRAP shows NIPBL chromatin residence time seems to increase a lot w/ PDS5 depletion

PDS5 inhibits cohesin-NIPBL ATPase+binding in vitro & suppresses NIPBL occupancy of cohesin in cells(+ PDS5&WAPL dont affect each others occupancy)
iFRAP shows NIPBL chromatin residence time seems to increase a lot w/ PDS5 depletion


Extrusion disrupts contacts. Compartmentalization results from equilibration via polymer relaxation
Fast cohesin turnover->disruptions occur more quickly than they can be relaxed


Extrusion disrupts contacts. Compartmentalization results from equilibration via polymer relaxation
Fast cohesin turnover->disruptions occur more quickly than they can be relaxed





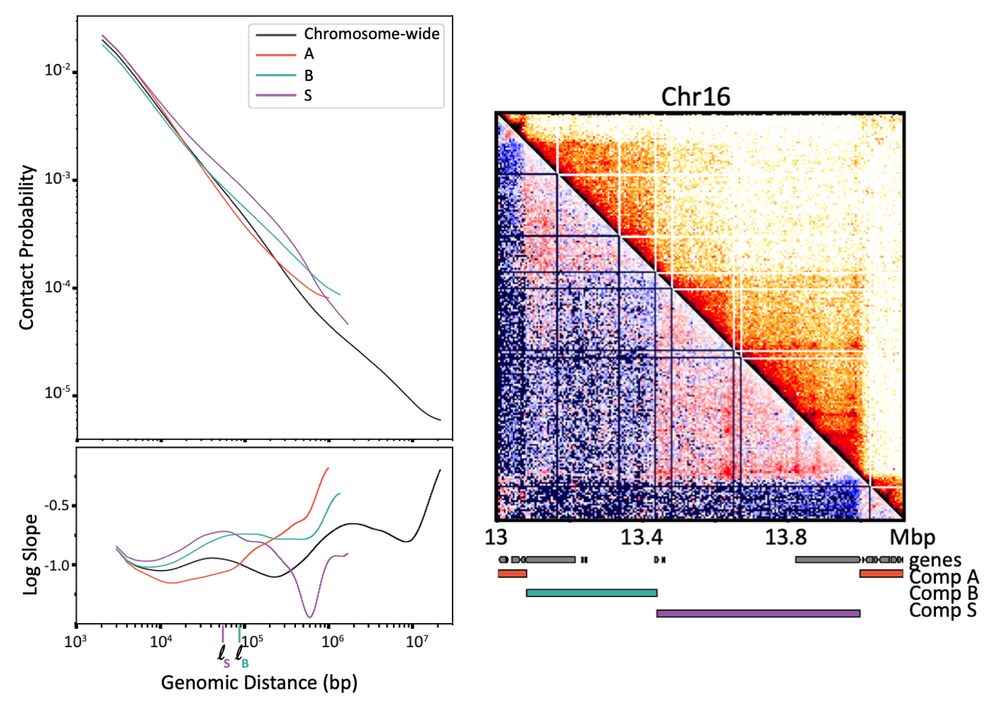
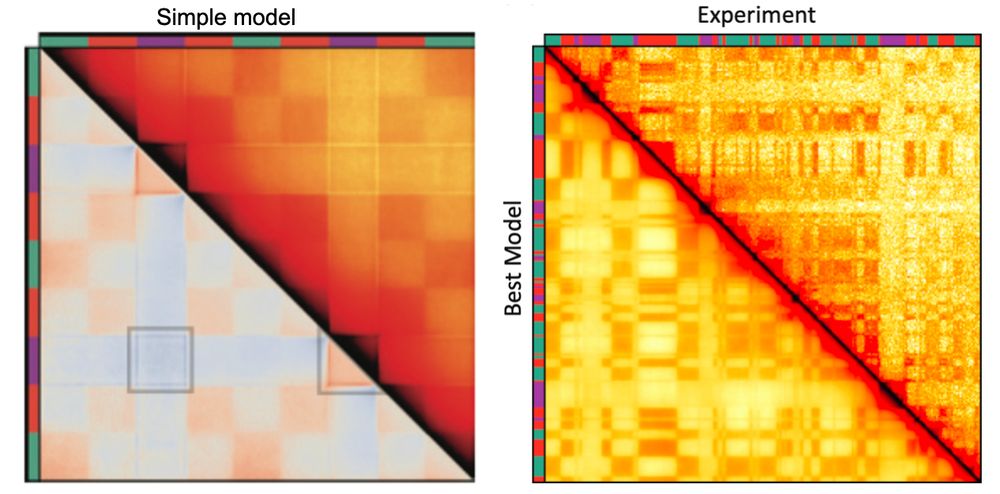
Emily’s simulations show S compartments can't be generated by either of these mechanisms alone! Phase separation makes unwanted inter-S contacts & extrusion can't deplete inter-S contacts w/out erasing all comps
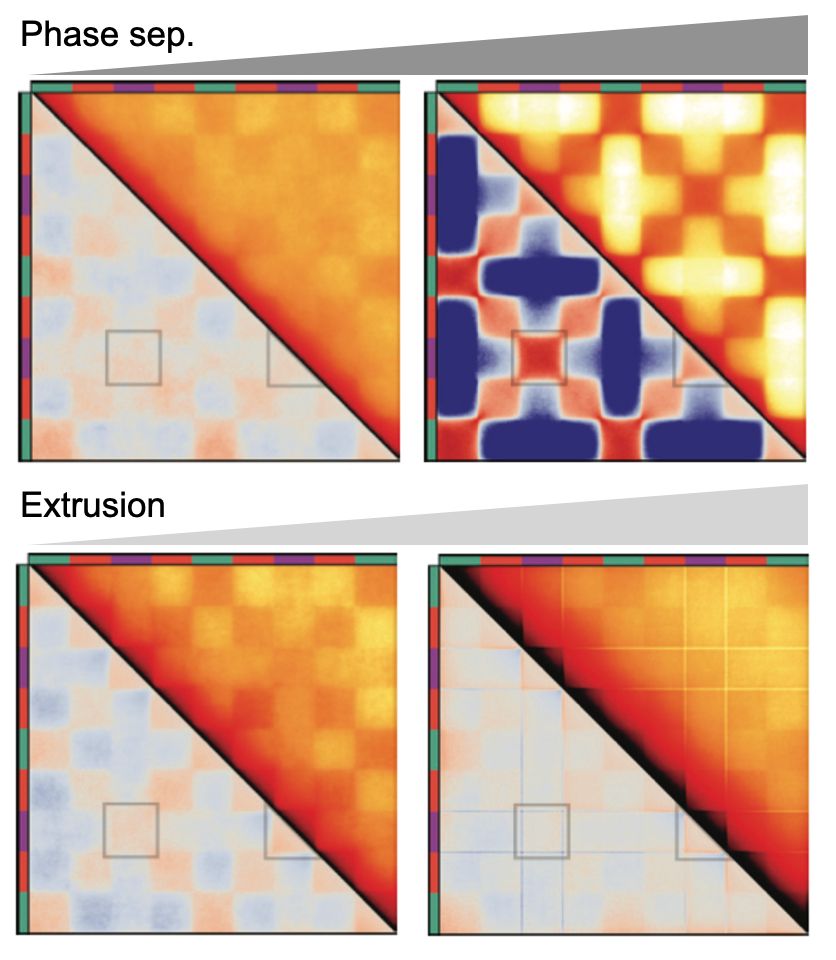
Emily’s simulations show S compartments can't be generated by either of these mechanisms alone! Phase separation makes unwanted inter-S contacts & extrusion can't deplete inter-S contacts w/out erasing all comps

More important, now, increasing phase-separated heterochromatin & crosslinked heterochromatin can increase nuclear stiffness.
Lamina tethers are physical constraints on chromatin!

More important, now, increasing phase-separated heterochromatin & crosslinked heterochromatin can increase nuclear stiffness.
Lamina tethers are physical constraints on chromatin!
Why? Chromatin condensation reduces polymer osmotic pressure. Stiffening heterochromatin by crosslinking doesnt help either! What’s going on?

Why? Chromatin condensation reduces polymer osmotic pressure. Stiffening heterochromatin by crosslinking doesnt help either! What’s going on?

1) affinity interactions
2) loop extrusion
3) chromatin volume density/compaction
Density seems to be particularly important & changes ~2-fold in M-to-G1 transition! But each factor seems to contribute to the observed dynamics

1) affinity interactions
2) loop extrusion
3) chromatin volume density/compaction
Density seems to be particularly important & changes ~2-fold in M-to-G1 transition! But each factor seems to contribute to the observed dynamics

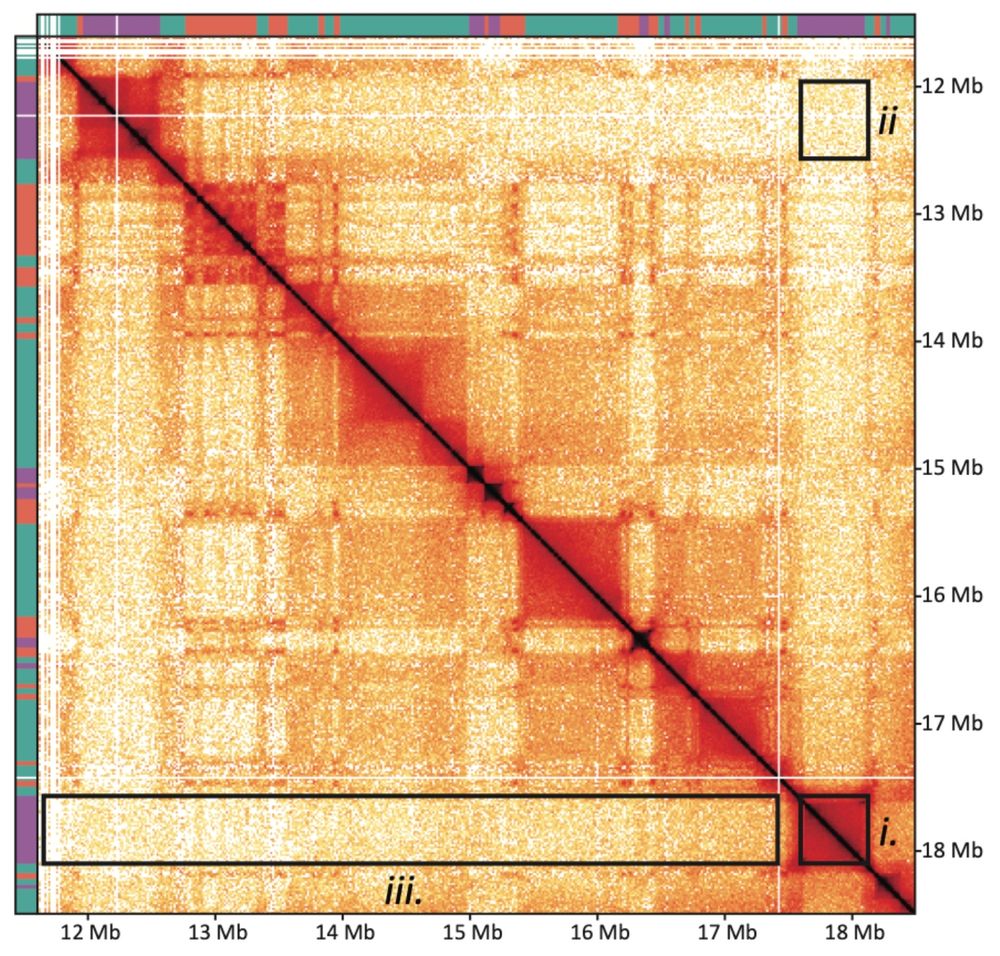
Using region capture Micro-C to deeply resolve we find chromosomes form microcompartments of cis-regulatory elements even during mitosis! They appear as a “grid of dots” in contact maps

Using region capture Micro-C to deeply resolve we find chromosomes form microcompartments of cis-regulatory elements even during mitosis! They appear as a “grid of dots” in contact maps


