







Tomorrow: We trace the ancient origins of this behavior and its impact on human disease.
Friday: Preprint drops!
Tomorrow: We trace the ancient origins of this behavior and its impact on human disease.
Friday: Preprint drops!
1) Make 2 genes w different signals, or
2) Decode a single RNA to make 2 proteins.
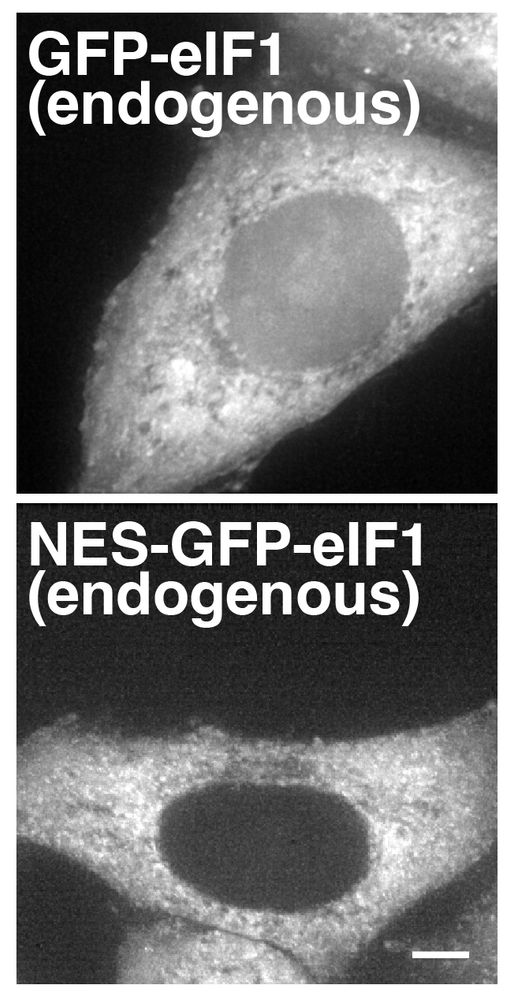
Our new work highlights how alternate translation initiation can create differentially localized proteins.
1) Make 2 genes w different signals, or
2) Decode a single RNA to make 2 proteins.
Our new work highlights how alternate translation initiation can create differentially localized proteins.
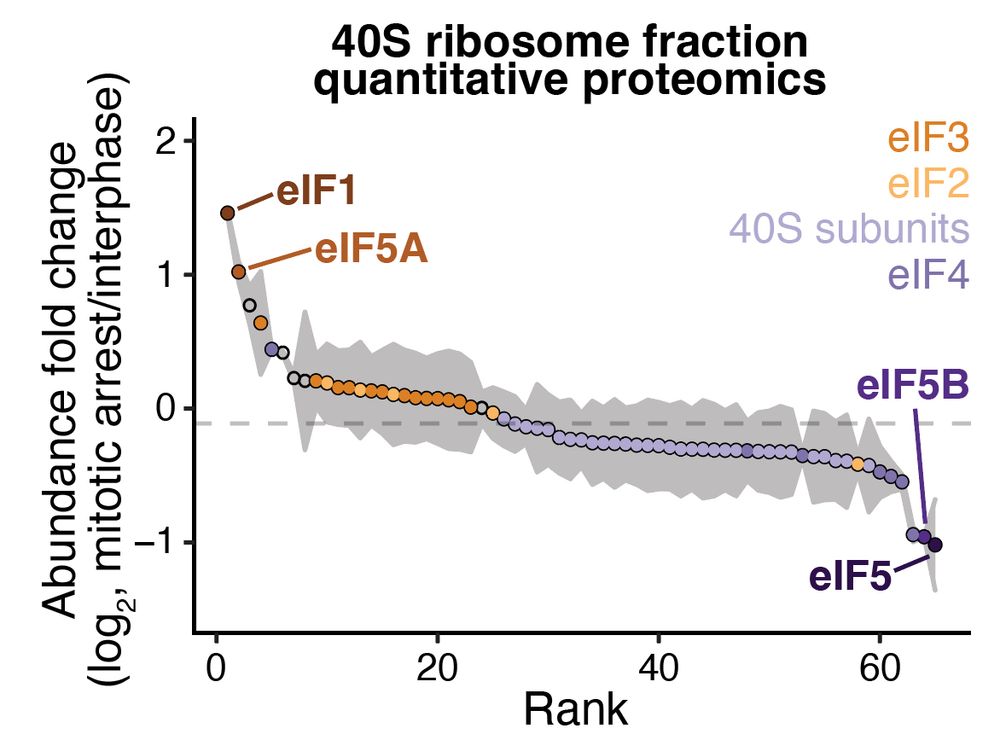
Mitochondria are cells within our cells. They need the same core activities - replication, transcription, translation. How do cells enable these diverse activities in both compartments? We uncover an unexpected + broad strategy with ancient origins. Stay tuned!
Mitochondria are cells within our cells. They need the same core activities - replication, transcription, translation. How do cells enable these diverse activities in both compartments? We uncover an unexpected + broad strategy with ancient origins. Stay tuned!

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
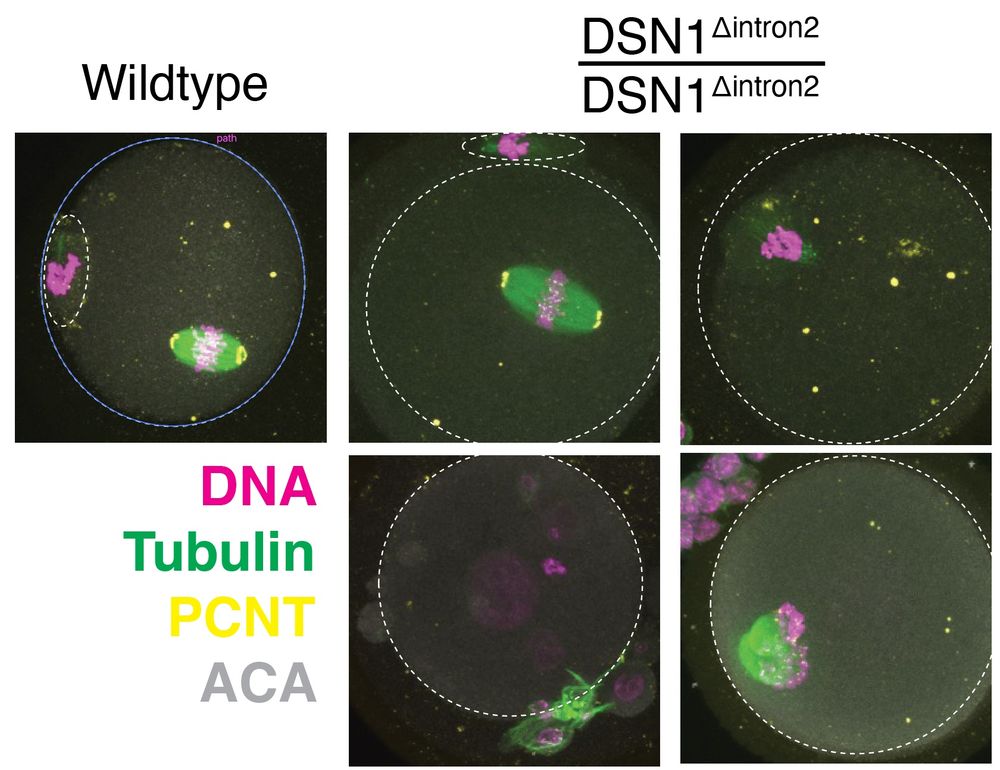
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...