Hugh Cottingham
@hughcottingham.bsky.social
Microbial genomics | Computational biology
Located at the Alfred Hospital/Monash Uni
Located at the Alfred Hospital/Monash Uni
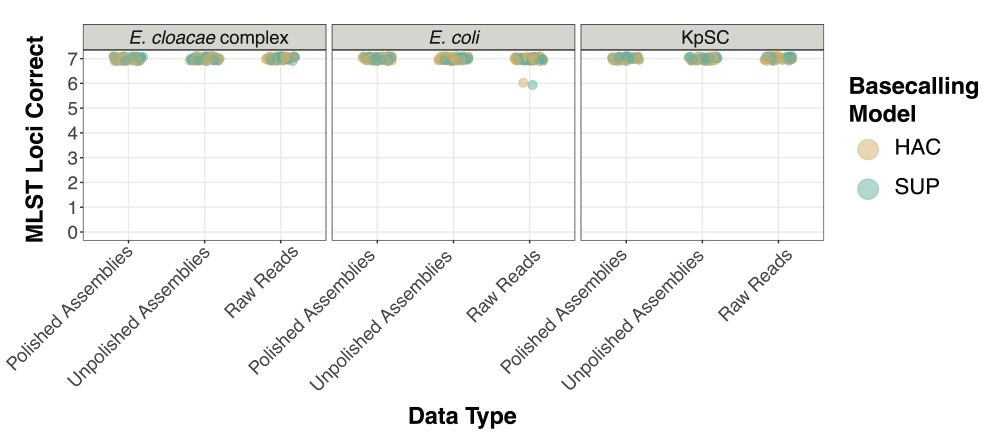
We found modern ONT data (R10.4.1, Dorado SUP) to generate perfect MLST profiles and AMR allelic variant profiles, ~99.5% cgMLST profiles and cgSNP profiles highly concordant with traditional Illumina methods /3
July 30, 2025 at 2:13 AM
We found modern ONT data (R10.4.1, Dorado SUP) to generate perfect MLST profiles and AMR allelic variant profiles, ~99.5% cgMLST profiles and cgSNP profiles highly concordant with traditional Illumina methods /3
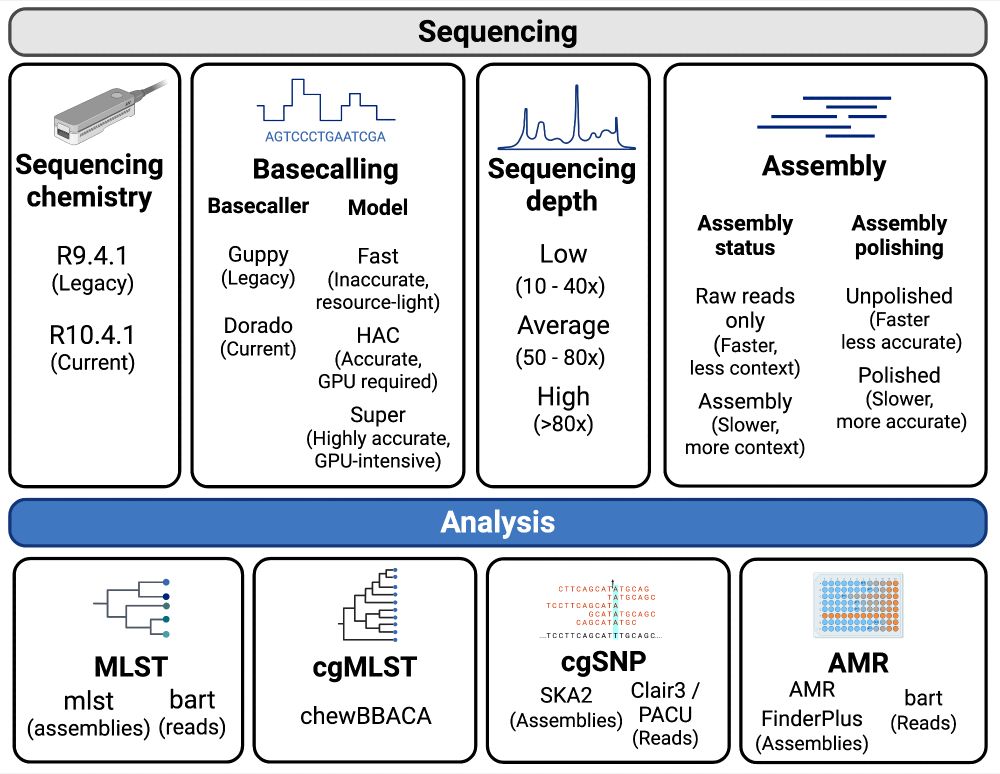
Nanopore data has come a long way the last few years, and offers users a huge amount of flexibility in how they want to generate data. We took all combinations of sequencing chemistry, basecalling, depth & assembly status from 3 Gram-negative species groups and compared to hybrid gold standards /2
July 30, 2025 at 2:12 AM
Nanopore data has come a long way the last few years, and offers users a huge amount of flexibility in how they want to generate data. We took all combinations of sequencing chemistry, basecalling, depth & assembly status from 3 Gram-negative species groups and compared to hybrid gold standards /2
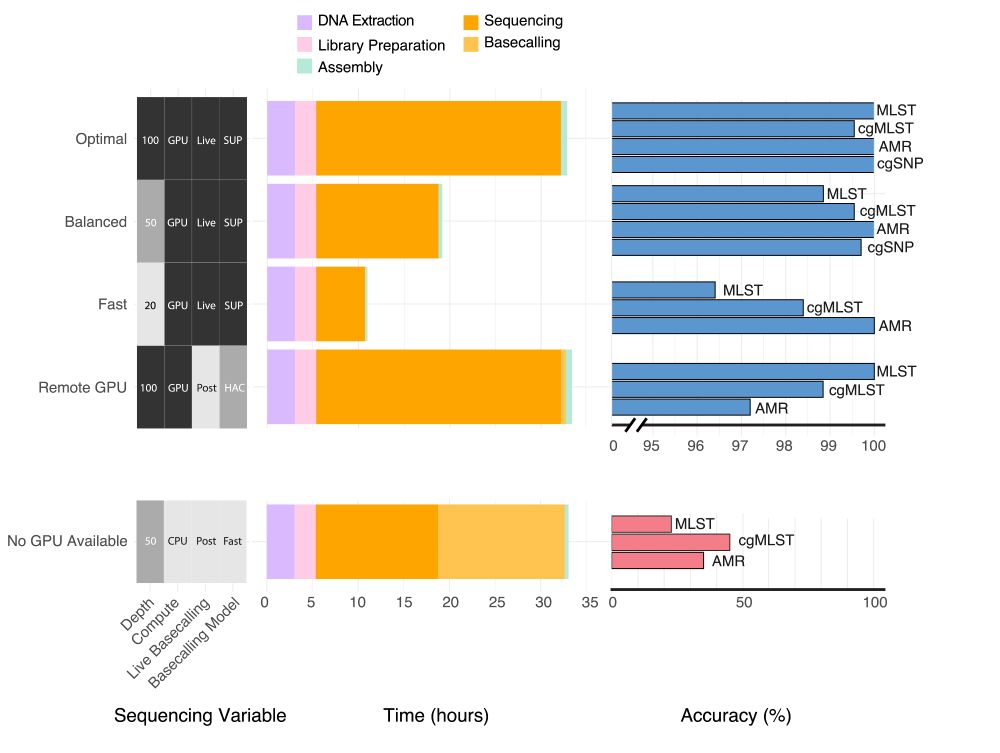
Pleased to say that our preprint benchmarking Nanopore data for MLST, cgMLST, cgSNP & AMR typing from bacterial isolates is out! TL;DR you can get almost perfect results from 50x depth using live SUP basecalling with a GPU in under 20 hours #microsky#IDsky 🦠🧬🖥️ /1
www.medrxiv.org/content/10.1...
www.medrxiv.org/content/10.1...
July 30, 2025 at 2:11 AM
Pleased to say that our preprint benchmarking Nanopore data for MLST, cgMLST, cgSNP & AMR typing from bacterial isolates is out! TL;DR you can get almost perfect results from 50x depth using live SUP basecalling with a GPU in under 20 hours #microsky#IDsky 🦠🧬🖥️ /1
www.medrxiv.org/content/10.1...
www.medrxiv.org/content/10.1...

