🌐 evedesign.bio
🌐 evedesign.bio
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
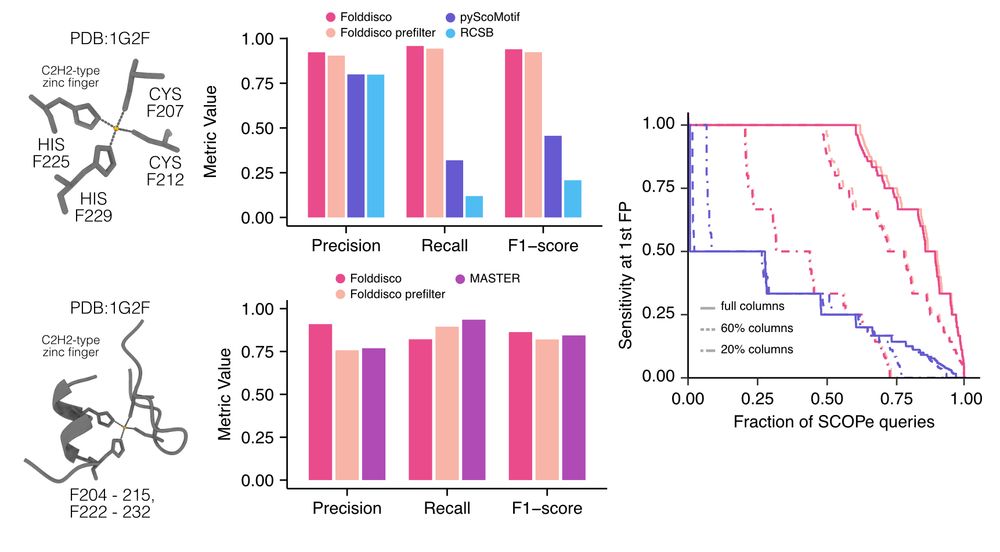
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
- Added description texts for AFDB
- Integrated TaxoView taxonomy visualization & filter by @sunjaelee.bsky.social
- Inter-residue distance clustering by DBSCAN to explore motif diversity.
🌐 search.foldseek.com/folddisco
📄 www.biorxiv.org/content/10.1...
📄 www.biorxiv.org/content/10.1...
Also check out poster:
B-50 lolalign Sensitive structural alignments by Lasse
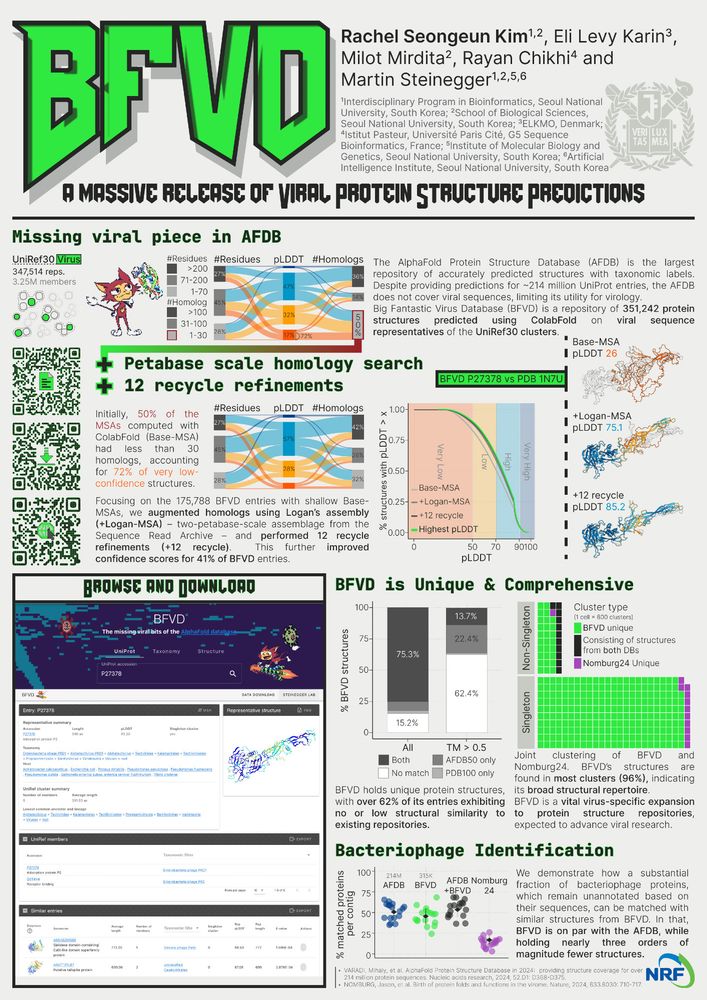
B-123 BFVD by Rachel

📄 www.biorxiv.org/content/10.1...
Also check out poster:
B-50 lolalign Sensitive structural alignments by Lasse
B-123 BFVD by Rachel
www.biorxiv.org/content/10.1...
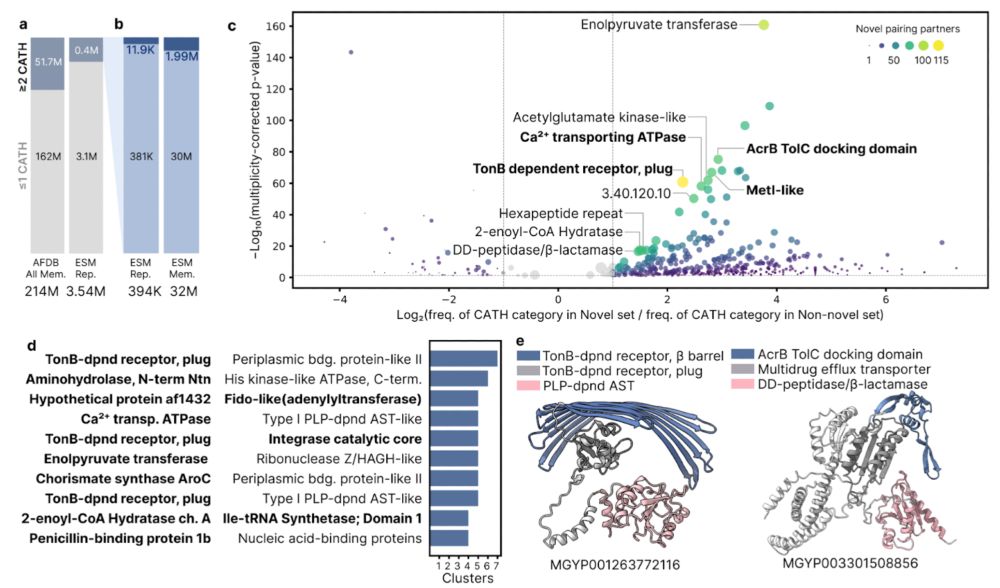
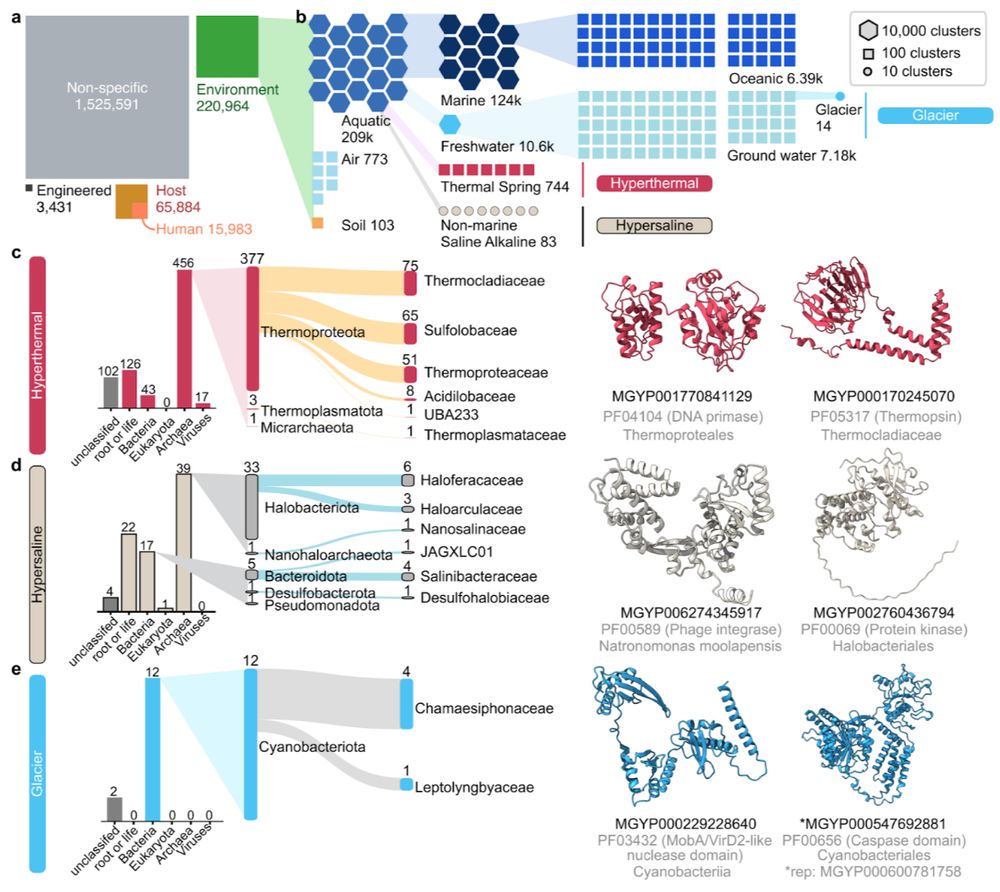
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social

www.biorxiv.org/content/10.1...
In this study, we present the largest systematic analysis of microbiome structure and function, integrating 85K uniformly processed metagenomes from diverse habitats worldwide.
@podlesny.bsky.social @jonas-bio.bsky.social @borklab.bsky.social
📄 academic.oup.com/nar/article/...

📄 academic.oup.com/nar/article/...
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
Read more about the work in the skeetorial
🦋 bsky.app/profile/mart...
or our preprint
📄 www.biorxiv.org/content/10.1...
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
Visit our #RECOMB2025 poster (719) & talk (1 pm at B145 on April 29).
Visit our #RECOMB2025 poster (719) & talk (1 pm at B145 on April 29).
Structural: MSAs, Virus DB, Core Genes, Motif Discovery, Multimer Clustering & Search, pLM Foldseek, Environmental analysis
Metagenomics: Classification & Metabuli App
GPU-based & RNA search, Proteome clustering, Novel Ribozyme discovery
& get Marv stickers!