








On zoom at 9am PT / 12pm ET / 6pm CE(S)T: portal.valencelabs.com/starklyspeak...

On zoom at 9am PT / 12pm ET / 6pm CE(S)T: portal.valencelabs.com/starklyspeak...
Experiments on the new OMol25 dataset!
On zoom 9am PT / noon ET / 6pm CEST: portal.valencelabs.com/starklyspeak...

Experiments on the new OMol25 dataset!
On zoom 9am PT / noon ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
We will also discuss how to close the gap between AF3 and its open source replications!
portal.valencelabs.com/starklyspeak...

We will also discuss how to close the gap between AF3 and its open source replications!
portal.valencelabs.com/starklyspeak...
arxiv.org/abs/2504.11713
Join us on zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
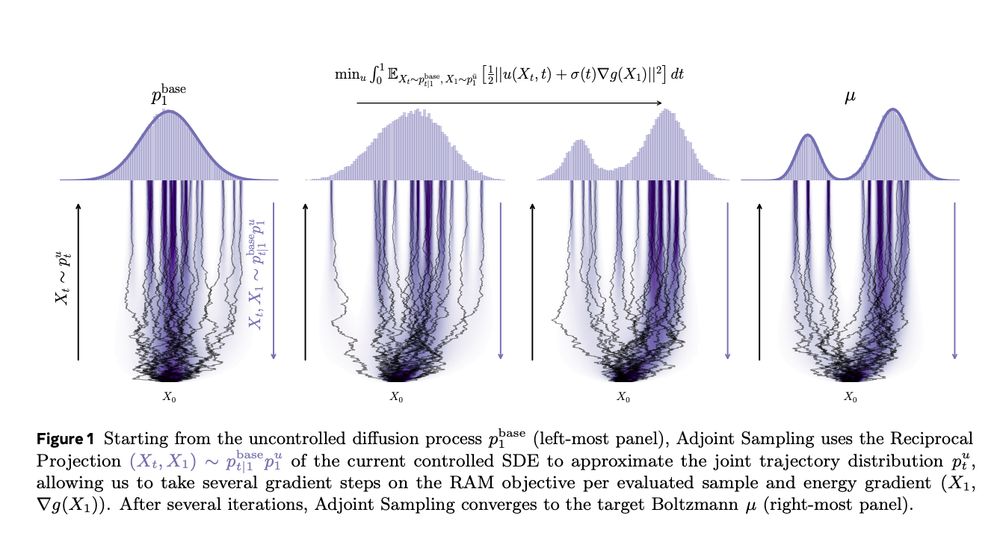
arxiv.org/abs/2504.11713
Join us on zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
Join us on zoom at 9am PT / 12pm ET: portal.valencelabs.com/starklyspeak...
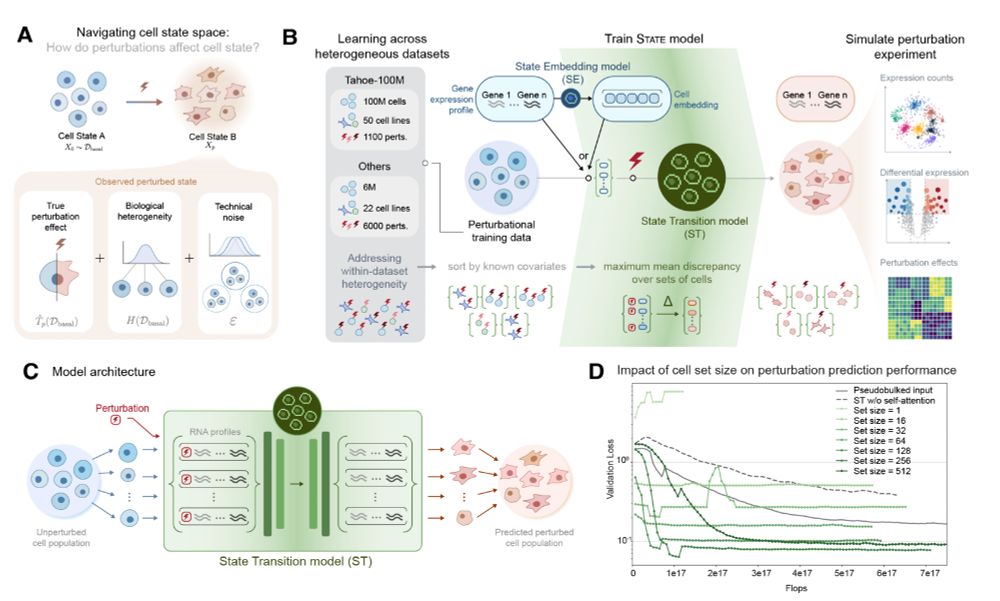
Join us on zoom at 9am PT / 12pm ET: portal.valencelabs.com/starklyspeak...
Join us on Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
Join us on Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
Via "Classifier-Free Guidance: From High-Dimensional Analysis to Generalized Guidance Forms" arxiv.org/abs/2502.07849
On Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...

Via "Classifier-Free Guidance: From High-Dimensional Analysis to Generalized Guidance Forms" arxiv.org/abs/2502.07849
On Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
bit.ly/boltz2-pdf
Structure models and careful affinity data handling result in an affinity predictor approaching FEP accuracy at 1000x the speed.
Zoom link: portal.valencelabs.com/starklyspeak...
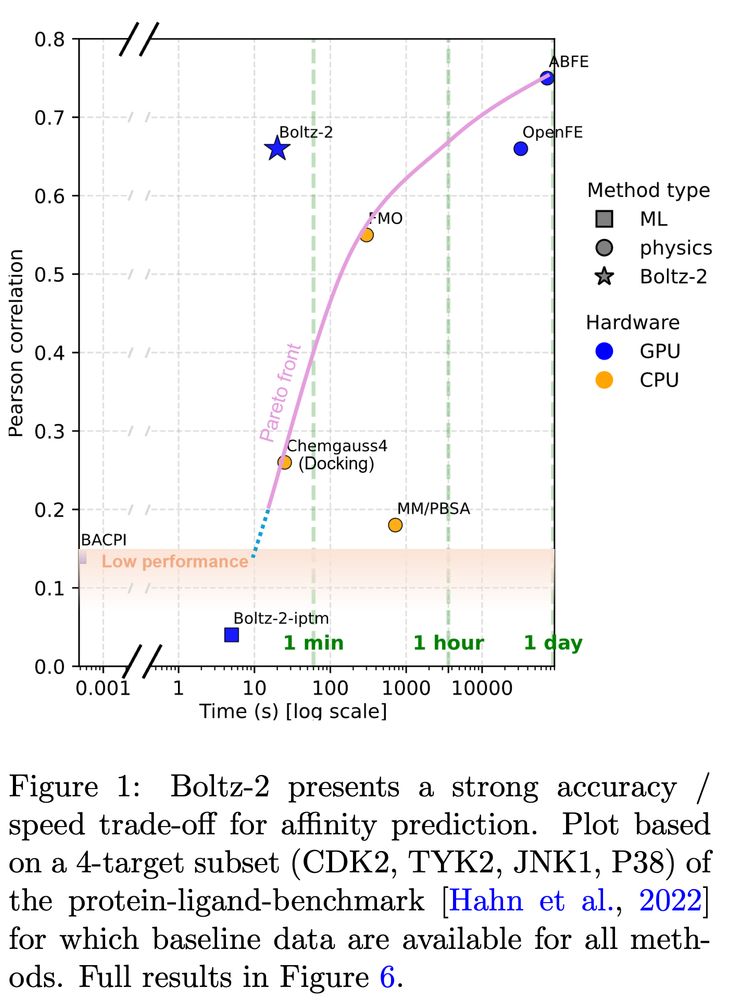
bit.ly/boltz2-pdf
Structure models and careful affinity data handling result in an affinity predictor approaching FEP accuracy at 1000x the speed.
Zoom link: portal.valencelabs.com/starklyspeak...
On zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
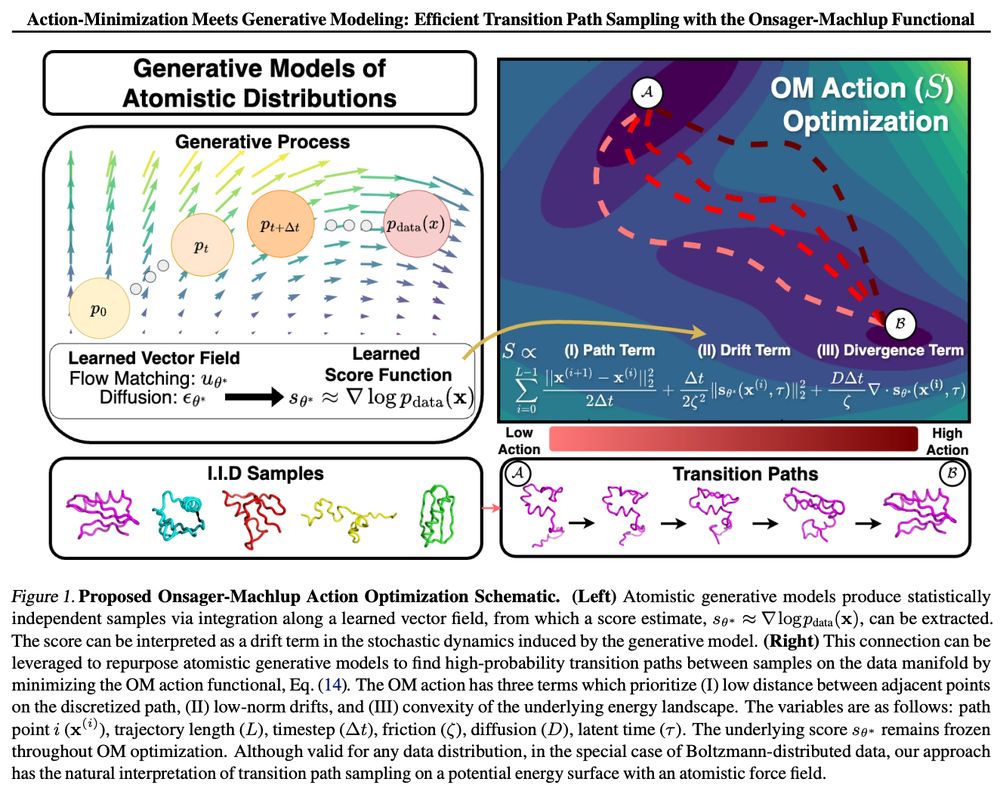
On zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
With @menghua_wu!
Join us on Zoom at 9am PT 12pm ET 6pm CEST: portal.valencelabs.com/starklyspeak...
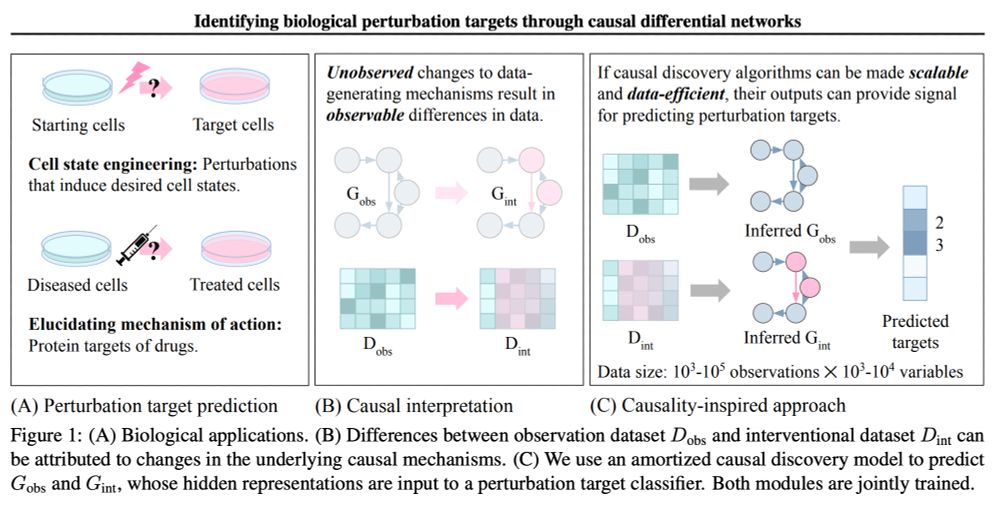
With @menghua_wu!
Join us on Zoom at 9am PT 12pm ET 6pm CEST: portal.valencelabs.com/starklyspeak...
On Zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
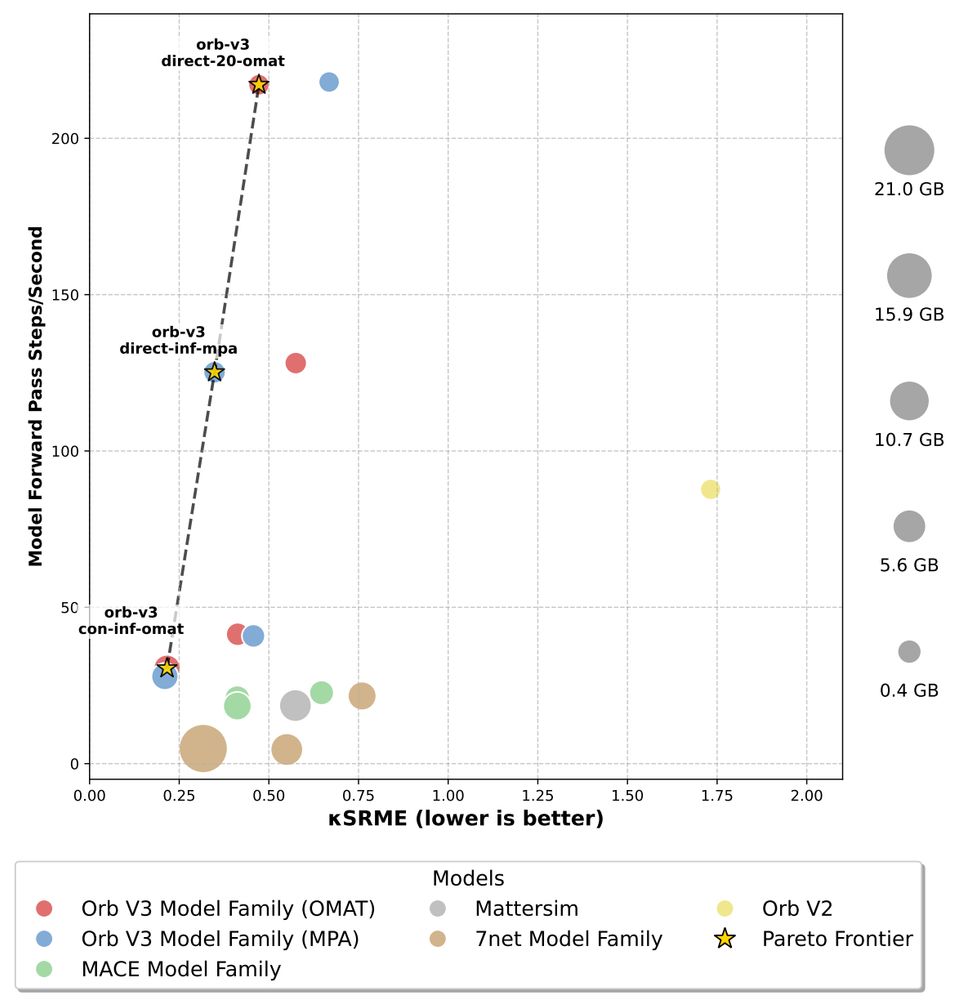
On Zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
Estimating FED is a strong tool for comparing drug's binding affinities
Join us on zoom at 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
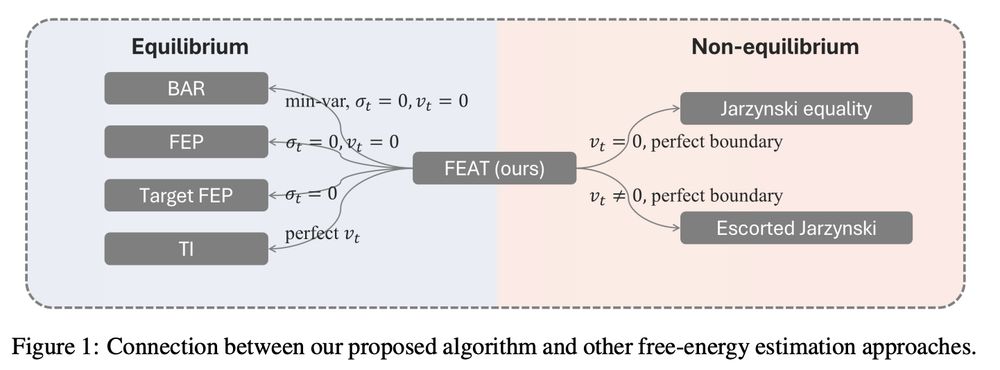
Estimating FED is a strong tool for comparing drug's binding affinities
Join us on zoom at 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
Join on Zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
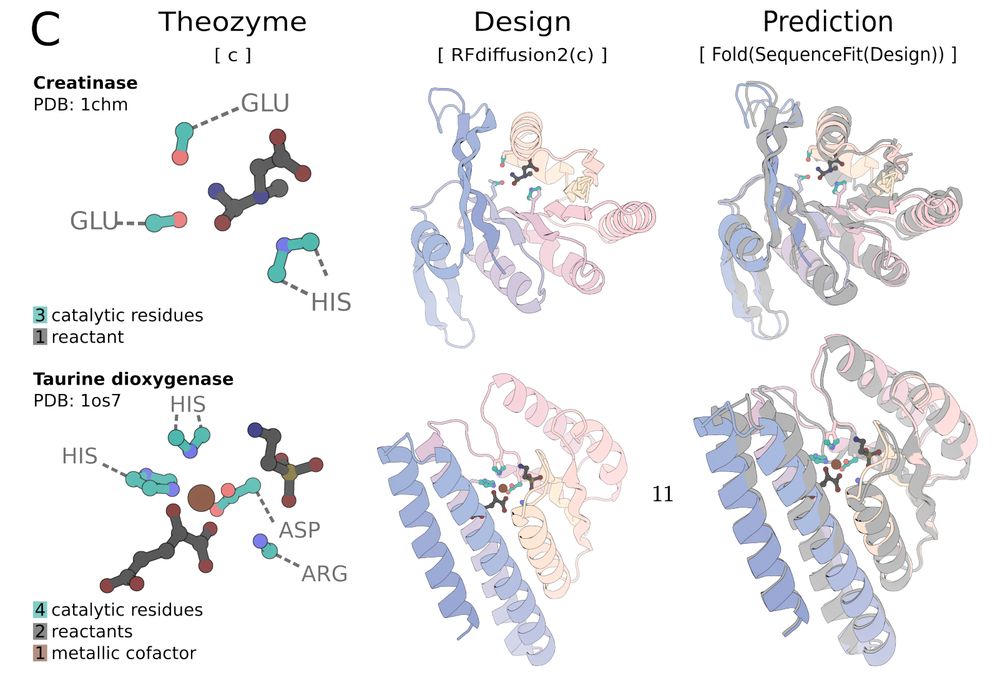
Join on Zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
On zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
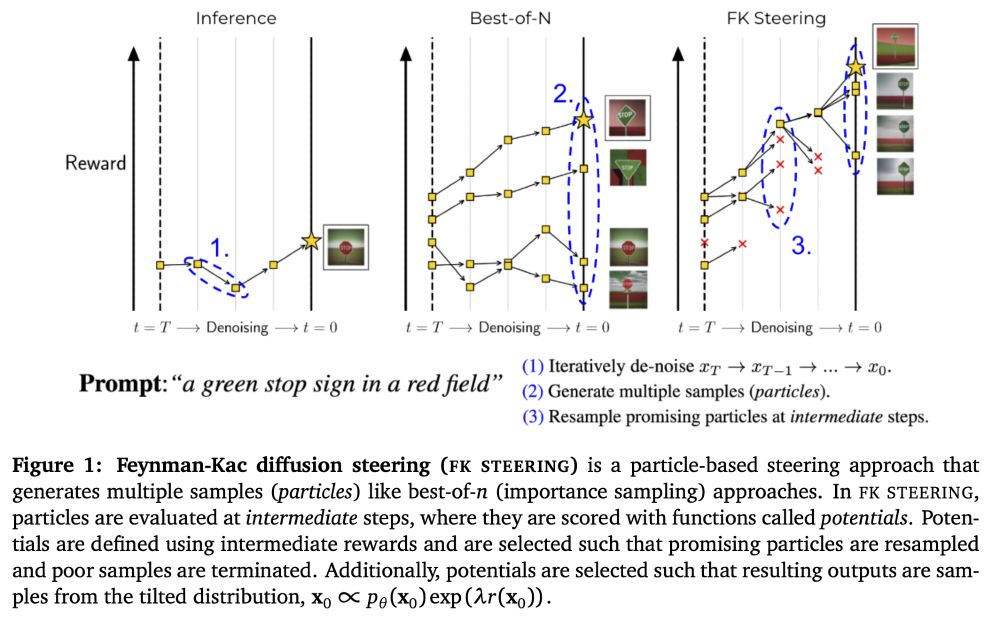
On zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
"One-step Diffusion Models with f-Divergence Distribution Matching" arxiv.org/abs/2502.15681
Presented by @xuyilun2
Zoom link: portal.valencelabs.com/logg

"One-step Diffusion Models with f-Divergence Distribution Matching" arxiv.org/abs/2502.15681
Presented by @xuyilun2
Zoom link: portal.valencelabs.com/logg
Humans:
Moi*, Bowen Jing*, Tomas Geffner, @jyim.bsky.social , Tommi Jaakkola, Arash Vahdat, @karstenkreis.bsky.social
Many of us will be at #ICLR2025 with this stuff, happy to chat!
Also happy to improve the NVIDIA logo a bit with this beauty made by Bowen🙃
6/6

Humans:
Moi*, Bowen Jing*, Tomas Geffner, @jyim.bsky.social , Tommi Jaakkola, Arash Vahdat, @karstenkreis.bsky.social
Many of us will be at #ICLR2025 with this stuff, happy to chat!
Also happy to improve the NVIDIA logo a bit with this beauty made by Bowen🙃
6/6

