


x.com/grkeele/status…

x.com/grkeele/status…



Using simulations from real genetic data, I sought to provide some answers and guidelines.
x.com/biorxivpreprin…

Using simulations from real genetic data, I sought to provide some answers and guidelines.
x.com/biorxivpreprin…
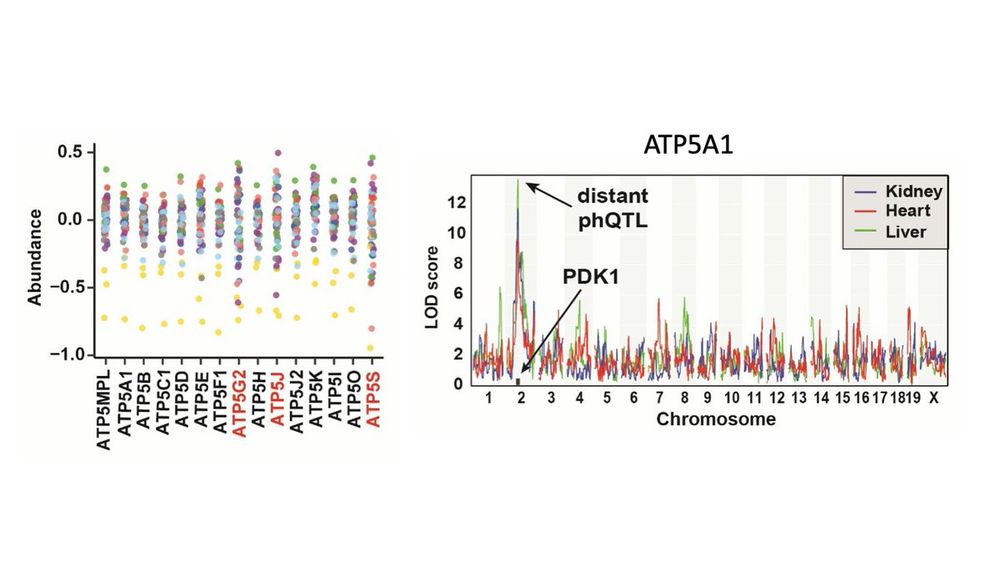



Here, we integrate expression, proteins, and phosphopeptides from three tissues of 58 inbred CC mouse strains.
doi.org/10.1016/j.xgen…

Here, we integrate expression, proteins, and phosphopeptides from three tissues of 58 inbred CC mouse strains.
doi.org/10.1016/j.xgen…



We compared to transcript data from a separate study of a sister strain of B6. This matches our previous findings in DO mice.

We compared to transcript data from a separate study of a sister strain of B6. This matches our previous findings in DO mice.









