
Gräter lab
@graeterlab.bsky.social
We aim at unraveling the intricacies of molecular perception and designing sensing molecular systems from the ground up. We work at the interface of biophysics, biochemistry, and materials science.
@ Max Planck Institute for Polymer Research, Mainz.
@ Max Planck Institute for Polymer Research, Mainz.
Reposted by Gräter lab
The #mattertolife fall days are a wonderful opportunity to connect to this vibrant community. This time at the @mpip-mainz.mpg.de with talks by the local hosts @graeterlab.bsky.social, @landfesterdept.bsky.social and @weilgroup.bsky.social. Thanks @mattertolife.bsky.social for bringing us here 👍😀🙏

September 22, 2025 at 4:14 PM
The #mattertolife fall days are a wonderful opportunity to connect to this vibrant community. This time at the @mpip-mainz.mpg.de with talks by the local hosts @graeterlab.bsky.social, @landfesterdept.bsky.social and @weilgroup.bsky.social. Thanks @mattertolife.bsky.social for bringing us here 👍😀🙏
Reposted by Gräter lab
Our #mattertolife Fall Days have just started 👏🏻 Thank you @mpip-mainz.mpg.de for hosting us!
@graeterlab.bsky.social @landfesterdept.bsky.social @weilgroup.bsky.social
@graeterlab.bsky.social @landfesterdept.bsky.social @weilgroup.bsky.social

September 22, 2025 at 7:13 AM
Our #mattertolife Fall Days have just started 👏🏻 Thank you @mpip-mainz.mpg.de for hosting us!
@graeterlab.bsky.social @landfesterdept.bsky.social @weilgroup.bsky.social
@graeterlab.bsky.social @landfesterdept.bsky.social @weilgroup.bsky.social
Reposted by Gräter lab
Flabby, flexible, and full of potential: Researchers at HITS and @mpip-mainz.mpg.de have developed a #machinelearning model that designs flexible proteins – even with rare patterns. A leap toward next-gen enzymes, therapeutics & sustainable biotech! 🌱💊🔬
👉 Read more: www.h-its.org/2025/07/23/f...
👉 Read more: www.h-its.org/2025/07/23/f...
July 23, 2025 at 12:47 PM
Flabby, flexible, and full of potential: Researchers at HITS and @mpip-mainz.mpg.de have developed a #machinelearning model that designs flexible proteins – even with rare patterns. A leap toward next-gen enzymes, therapeutics & sustainable biotech! 🌱💊🔬
👉 Read more: www.h-its.org/2025/07/23/f...
👉 Read more: www.h-its.org/2025/07/23/f...
Want your de novo designed protein to be wobbly? 🍮 Seva, Leif & team have made it possible - you find them at ICML! 🤩 @mpip-mainz.mpg.de @hitsters.bsky.social
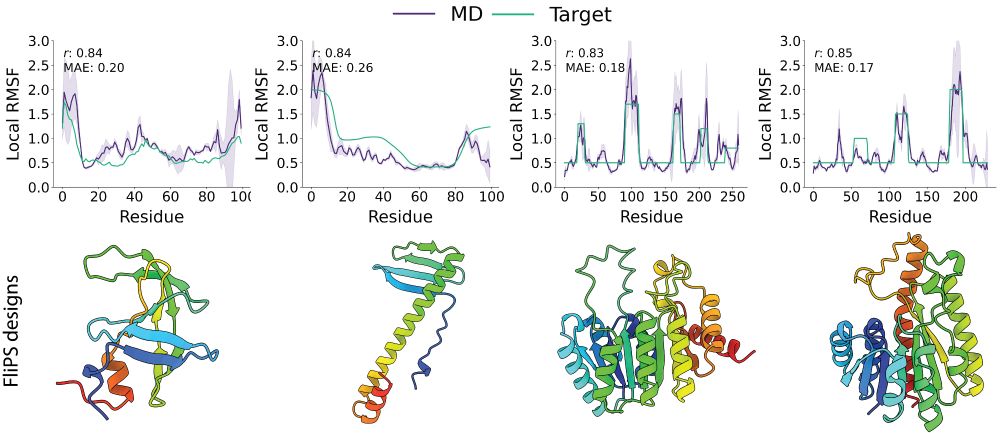
1/6 One of the key features of functional proteins is their inherent structural flexibility. In our recent work at #ICML, we introduce flexibility to protein structure design! More in a thread below.
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
Code / Tutorial: github.com/graeter-grou...
Poster: W-109, Thu 17 Jul 11 a.m. PDT — 1:30 p.m. PDT
July 17, 2025 at 6:29 PM
Want your de novo designed protein to be wobbly? 🍮 Seva, Leif & team have made it possible - you find them at ICML! 🤩 @mpip-mainz.mpg.de @hitsters.bsky.social
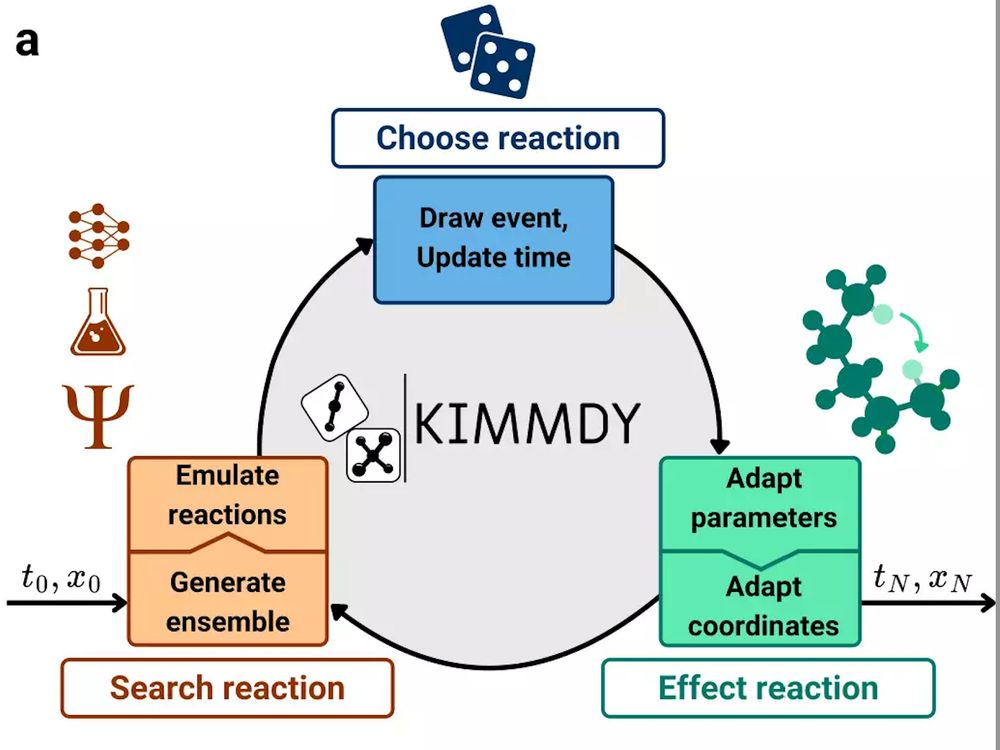
You want to make your simulations reactive, at little cost? Try KIMMDY. Don't hesitate to reach out if you want to include your favorite chemistry. Super happy to see this online, kudos to the team!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
July 7, 2025 at 7:00 AM
You want to make your simulations reactive, at little cost? Try KIMMDY. Don't hesitate to reach out if you want to include your favorite chemistry. Super happy to see this online, kudos to the team!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Gräter lab
I thought GRAPPA would also make a nice name (going with the MARTINI, CALVADOS etc... trend) and it turns out it already exists ! It's a model from the @graeterlab.bsky.social :
pubs.rsc.org/en/content/a...
pubs.rsc.org/en/content/a...

Grappa – a machine learned molecular mechanics force field
Simulating large molecular systems over long timescales requires force fields that are both accurate and efficient. In recent years, E(3) equivariant neural networks have lifted the tension between computational efficiency and accuracy of force fields, but they are still several orders of magnitude more expe
pubs.rsc.org
July 1, 2025 at 7:10 AM
I thought GRAPPA would also make a nice name (going with the MARTINI, CALVADOS etc... trend) and it turns out it already exists ! It's a model from the @graeterlab.bsky.social :
pubs.rsc.org/en/content/a...
pubs.rsc.org/en/content/a...
Join us at the MPIP in Mainz - a few days left to apply! You feel this matches your research agenda but would be a step too early in your career? Do apply!
Job alert: Join us in Mainz as
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de

Max Planck Research Group Leader (W2) in Molecular Design
We are looking for exceptional early-career scientists conducting computational research with a proven record of accomplishment. The primary focus of this call is on candidates proposing research on b...
tinyurl.com
May 9, 2025 at 5:30 AM
Join us at the MPIP in Mainz - a few days left to apply! You feel this matches your research agenda but would be a step too early in your career? Do apply!
Reposted by Gräter lab
@rebecca-wade.bsky.social @graeterlab.bsky.social @janstuehmer.bsky.social @astroinformatics.bsky.social @leif-seute.bsky.social
The 3-day SIMPLAIX Workshop 2025 is now officially OPEN! 🎉
We’re kicking off an inspiring event filled with keynotes, discussions and networking opportunities on multiscale computer simulation and #machinelearning over the next few days.
#SIMPLAIX2025 #AI #lifesciences #materialsscience
We’re kicking off an inspiring event filled with keynotes, discussions and networking opportunities on multiscale computer simulation and #machinelearning over the next few days.
#SIMPLAIX2025 #AI #lifesciences #materialsscience




May 7, 2025 at 1:00 PM
Reposted by Gräter lab
Registration Deadline Extended!
The SIMPLAIX Workshop 2025 is just around the corner, and there's still time to join us!
📅 New deadline: 13 April 2025
📍 Studio Villa Bosch, Heidelberg
🔗 Register now: simplaix-workshop2025.h-its.org
Spots are filling fast — secure yours today! #SIMPLAIX25
The SIMPLAIX Workshop 2025 is just around the corner, and there's still time to join us!
📅 New deadline: 13 April 2025
📍 Studio Villa Bosch, Heidelberg
🔗 Register now: simplaix-workshop2025.h-its.org
Spots are filling fast — secure yours today! #SIMPLAIX25

April 7, 2025 at 12:50 PM
Registration Deadline Extended!
The SIMPLAIX Workshop 2025 is just around the corner, and there's still time to join us!
📅 New deadline: 13 April 2025
📍 Studio Villa Bosch, Heidelberg
🔗 Register now: simplaix-workshop2025.h-its.org
Spots are filling fast — secure yours today! #SIMPLAIX25
The SIMPLAIX Workshop 2025 is just around the corner, and there's still time to join us!
📅 New deadline: 13 April 2025
📍 Studio Villa Bosch, Heidelberg
🔗 Register now: simplaix-workshop2025.h-its.org
Spots are filling fast — secure yours today! #SIMPLAIX25
Kai's work among 'Most popular articles 2024' in Chem Sci - many thanks to the readers :-)
tinyurl.com/4yb78j5e
Link to our work to predict energy barriers from reactants without DFT: tinyurl.com/5cm4h3ka
@hitsters.bsky.social @mpip-mainz.mpg.de
tinyurl.com/4yb78j5e
Link to our work to predict energy barriers from reactants without DFT: tinyurl.com/5cm4h3ka
@hitsters.bsky.social @mpip-mainz.mpg.de
April 8, 2025 at 11:51 AM
Kai's work among 'Most popular articles 2024' in Chem Sci - many thanks to the readers :-)
tinyurl.com/4yb78j5e
Link to our work to predict energy barriers from reactants without DFT: tinyurl.com/5cm4h3ka
@hitsters.bsky.social @mpip-mainz.mpg.de
tinyurl.com/4yb78j5e
Link to our work to predict energy barriers from reactants without DFT: tinyurl.com/5cm4h3ka
@hitsters.bsky.social @mpip-mainz.mpg.de
Job alert: Join us in Mainz as
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de

Max Planck Research Group Leader (W2) in Molecular Design
We are looking for exceptional early-career scientists conducting computational research with a proven record of accomplishment. The primary focus of this call is on candidates proposing research on b...
tinyurl.com
April 4, 2025 at 4:00 PM
Job alert: Join us in Mainz as
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de
A while online now already: Predict the barrier of a reaction without knowing the transition state and in the low data regime using Gaussian Process regr.
Led by Evgeni Ulanov, with Ghulam, Kai and Pascal Friederich @ KIT.
@mpip-mainz.mpg.de @hitsters.bsky.social
pubs.rsc.org/en/content/a...
Led by Evgeni Ulanov, with Ghulam, Kai and Pascal Friederich @ KIT.
@mpip-mainz.mpg.de @hitsters.bsky.social
pubs.rsc.org/en/content/a...
March 31, 2025 at 12:04 PM
A while online now already: Predict the barrier of a reaction without knowing the transition state and in the low data regime using Gaussian Process regr.
Led by Evgeni Ulanov, with Ghulam, Kai and Pascal Friederich @ KIT.
@mpip-mainz.mpg.de @hitsters.bsky.social
pubs.rsc.org/en/content/a...
Led by Evgeni Ulanov, with Ghulam, Kai and Pascal Friederich @ KIT.
@mpip-mainz.mpg.de @hitsters.bsky.social
pubs.rsc.org/en/content/a...
Reposted by Gräter lab
Would you present your next NeurIPS paper in Europe instead of traveling to San Diego (US) if this was an option? Søren Hauberg (DTU) and I would love to hear the answer through this poll: (1/6)

NeurIPS participation in Europe
We seek to understand if there is interest in being able to attend NeurIPS in Europe, i.e. without travelling to San Diego, US. In the following, assume that it is possible to present accepted papers ...
docs.google.com
March 30, 2025 at 6:04 PM
Would you present your next NeurIPS paper in Europe instead of traveling to San Diego (US) if this was an option? Søren Hauberg (DTU) and I would love to hear the answer through this poll: (1/6)
Wondering how to predict protein flexibility in a sec? No time to run MD simulations but want to go beyond pLDDT? Check out BBFlow
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
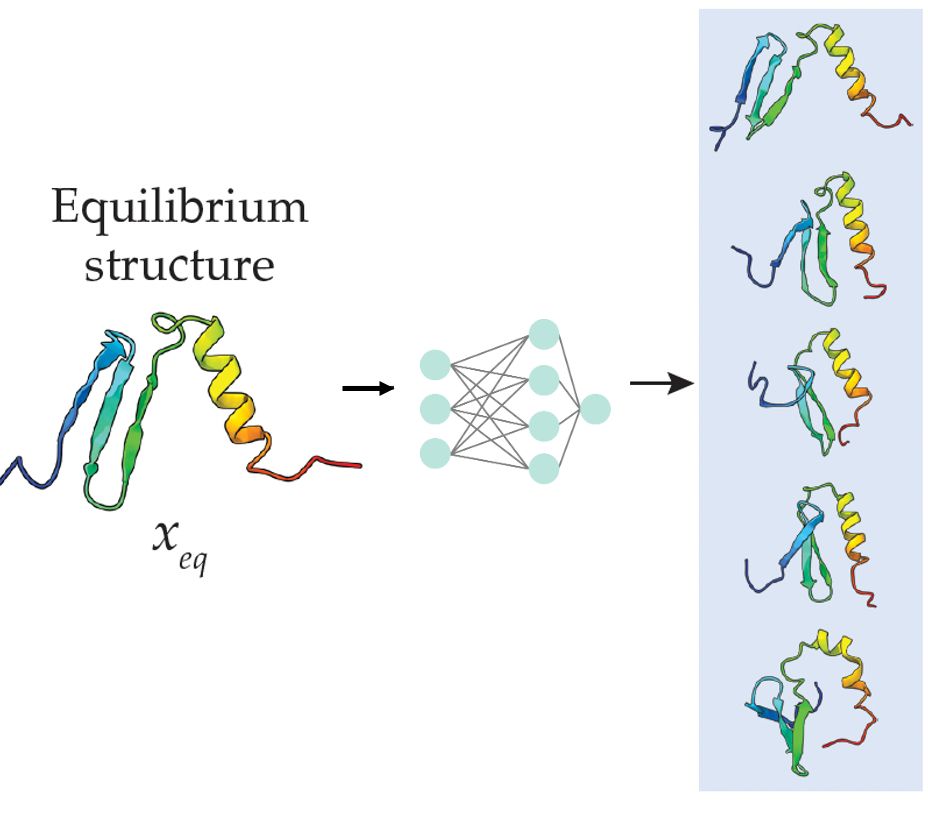
March 31, 2025 at 11:37 AM
Wondering how to predict protein flexibility in a sec? No time to run MD simulations but want to go beyond pLDDT? Check out BBFlow
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
arxiv.org/html/2503.05...
Useful in particular for de novo designs.
Led by Nico Wolf & Leif Seute, w Seva, Simon, and Jan. @mpip-mainz.mpg.de @hitsters.bsky.social
Reposted by Gräter lab
#compchem Good read: Crash testing machine learning force fields for molecules, materials, and interfaces: model analysis in the TEA Challenge 2023 #compchemsky doi.org/10.1039/D4SC...
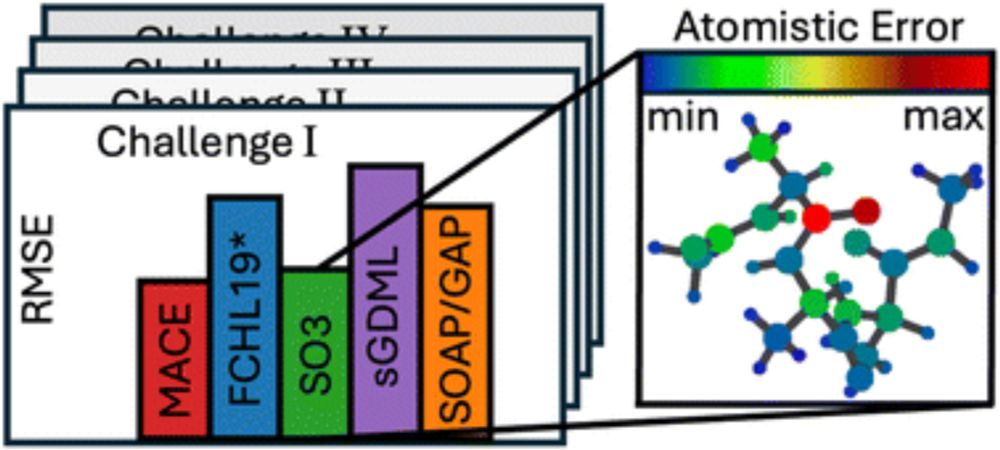
Crash testing machine learning force fields for molecules, materials, and interfaces: model analysis in the TEA Challenge 2023
Atomistic simulations are routinely employed in academia and industry to study the behavior of molecules, materials, and their interfaces. Central to these simulations are force fields (FFs), whose de...
doi.org
February 12, 2025 at 9:21 AM
#compchem Good read: Crash testing machine learning force fields for molecules, materials, and interfaces: model analysis in the TEA Challenge 2023 #compchemsky doi.org/10.1039/D4SC...
Reposted by Gräter lab
Curious about how Machine Learning and Molecular Simulations are used to model Protein Dynamics?
Then watch our HITS-SIMPLAIX Joint Colloquium with Cecilia Clementi @cecclementi.bsky.social which is now available on our YouTube channel.
www.youtube.com/watch?v=2HAI...
Then watch our HITS-SIMPLAIX Joint Colloquium with Cecilia Clementi @cecclementi.bsky.social which is now available on our YouTube channel.
www.youtube.com/watch?v=2HAI...

HITS-SIMPLAIX Joint Colloquium: Cecilia Clementi on protein dynamics and Machine Learning
YouTube video by TheHITSters
www.youtube.com
February 12, 2025 at 1:57 PM
Curious about how Machine Learning and Molecular Simulations are used to model Protein Dynamics?
Then watch our HITS-SIMPLAIX Joint Colloquium with Cecilia Clementi @cecclementi.bsky.social which is now available on our YouTube channel.
www.youtube.com/watch?v=2HAI...
Then watch our HITS-SIMPLAIX Joint Colloquium with Cecilia Clementi @cecclementi.bsky.social which is now available on our YouTube channel.
www.youtube.com/watch?v=2HAI...
Reposted by Gräter lab
📢 It is 5 min before 12❗
"Aufstehen für Demokratie" is committed to fact-based political decisions & democratic processes:
www.aufstehenfuerdemokratie.de
www.openpetition.de/!ychtb
📅 Join us on Feb 12th at 11:55 am at universities and research institutions in Germany.
🔬 Science needs democracy 🤝
"Aufstehen für Demokratie" is committed to fact-based political decisions & democratic processes:
www.aufstehenfuerdemokratie.de
www.openpetition.de/!ychtb
📅 Join us on Feb 12th at 11:55 am at universities and research institutions in Germany.
🔬 Science needs democracy 🤝

February 10, 2025 at 8:33 AM
Reposted by Gräter lab
Don't miss our next HITS-SIMPLAIX Joint Colloquium on 10 February at 2:00 PM CET! Cecilia Clementi @cecclementi.bsky.social will talk about Modeling Protein Dynamics with Machine Learning and Molecular Simulation. Sign up ow.ly/h0Wl50UQ2kX to join via Zoom or just come by. @simplaix.bsky.social
February 3, 2025 at 2:38 PM
Don't miss our next HITS-SIMPLAIX Joint Colloquium on 10 February at 2:00 PM CET! Cecilia Clementi @cecclementi.bsky.social will talk about Modeling Protein Dynamics with Machine Learning and Molecular Simulation. Sign up ow.ly/h0Wl50UQ2kX to join via Zoom or just come by. @simplaix.bsky.social
Reposted by Gräter lab
This was a very interesting paper to help supervise because it’s a totally different way to think about DNA structure, and uses a sequencer in a very creative way to understand DNA breakage. Check it out! 🧬
Where does DNA break when pulled? We can tune it! Paper led by Johannes Hahmann, MD simulations by Boris, great collaboration with Herrmann and Goestl labs @ RTWH
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
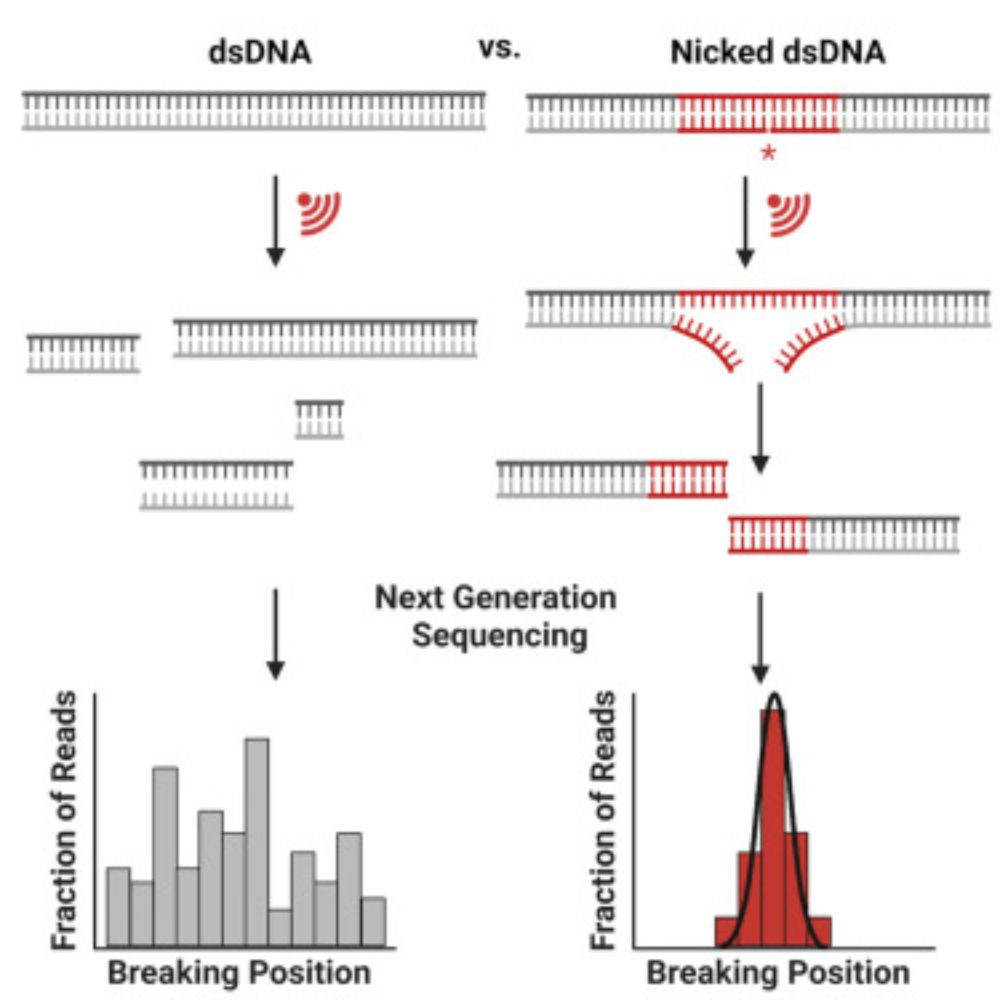
Sequence-specific, mechanophore-free mechanochemistry of DNA
Nucleic acids, such as DNA, are integral components of biological systems in that they steer many cellular processes and biotechnological applications…
www.sciencedirect.com
January 17, 2025 at 10:19 AM
This was a very interesting paper to help supervise because it’s a totally different way to think about DNA structure, and uses a sequencer in a very creative way to understand DNA breakage. Check it out! 🧬
Reposted by Gräter lab
Need forcefield parameters for a weird small molecule, modified nucleotide, or non-natural amino acid? Check out Grappa, a GNN approach to generating bonded parameters compatible with the MD engine of your choice! 🧬
Grappa is out: pubs.rsc.org/en/content/a...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...

Grappa – a machine learned molecular mechanics force field
Simulating large molecular systems over long timescales requires force fields that are both accurate and efficient. In recent years, E(3) equivariant neural networks have lifted the tension between co...
pubs.rsc.org
January 27, 2025 at 4:41 PM
Need forcefield parameters for a weird small molecule, modified nucleotide, or non-natural amino acid? Check out Grappa, a GNN approach to generating bonded parameters compatible with the MD engine of your choice! 🧬
Grappa is out: pubs.rsc.org/en/content/a...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...

Grappa – a machine learned molecular mechanics force field
Simulating large molecular systems over long timescales requires force fields that are both accurate and efficient. In recent years, E(3) equivariant neural networks have lifted the tension between co...
pubs.rsc.org
January 24, 2025 at 2:46 PM
Grappa is out: pubs.rsc.org/en/content/a...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...
We are looking forward to feedback from and extensions by the community! Try it out for your favorite (bio)molecules - ligands, post-translational modifications, metal-enzymes, DNA, ...
You find us here:
https://bsky.app/profile/graeterlab.bsky.social
https://bsky.app/profile/graeterlab.bsky.social

Gräter lab (@graeterlab.bsky.social)
We aim at unraveling the intricacies of molecular perception and designing sensing molecular systems from the ground up. We work at the interface of biophysics, biochemistry, and materials science.
@ Max Planck Institute for Polymer Research, Mainz.
bsky.app
January 27, 2025 at 8:40 AM
You find us here:
https://bsky.app/profile/graeterlab.bsky.social
https://bsky.app/profile/graeterlab.bsky.social
Reposted by Gräter lab
A long journey has taken the next step. Yesterday we defended our Cluster of Excellence Proposal #CoM2Life Communicating Biomaterials at the DFG. What an amazing journey with an amazing team. #teamscienceisthedreamscience @unimainz.bsky.social @tuda.bsky.social @mpip-mainz.mpg.de

January 15, 2025 at 2:49 PM
A long journey has taken the next step. Yesterday we defended our Cluster of Excellence Proposal #CoM2Life Communicating Biomaterials at the DFG. What an amazing journey with an amazing team. #teamscienceisthedreamscience @unimainz.bsky.social @tuda.bsky.social @mpip-mainz.mpg.de
Where does DNA break when pulled? We can tune it! Paper led by Johannes Hahmann, MD simulations by Boris, great collaboration with Herrmann and Goestl labs @ RTWH
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...

Sequence-specific, mechanophore-free mechanochemistry of DNA
Nucleic acids, such as DNA, are integral components of biological systems in that they steer many cellular processes and biotechnological applications…
www.sciencedirect.com
January 17, 2025 at 8:17 AM
Where does DNA break when pulled? We can tune it! Paper led by Johannes Hahmann, MD simulations by Boris, great collaboration with Herrmann and Goestl labs @ RTWH
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
🥲... and very excited to start from January with full forces at the MPIP@Mainz, with many (most) of my awesome lab members - and some HITS spirits - tagging along
After 15 years, we say goodbye to group leader Frauke Gräter (@graeterlab.bsky.social) who will assume her new role as director of the Max-Planck-Institute for Polymer Research in Mainz as of January 2025.
Read more about her time at HITS here: www.h-its.org/2024/12/19/f...
Read more about her time at HITS here: www.h-its.org/2024/12/19/f...

Frauke Gräter: “I will miss HITS.
After fifteen years at HITS, group leader Frauke Gräter will leave the institute at the end of the year. As ...
www.h-its.org
December 20, 2024 at 8:58 PM
🥲... and very excited to start from January with full forces at the MPIP@Mainz, with many (most) of my awesome lab members - and some HITS spirits - tagging along

