This tool allows users to estimate the genetic prevalence of autosomal recessive diseases using #gnomAD allele frequency data & classifications from #ClinVar
Blog post: broad.io/genie_blog
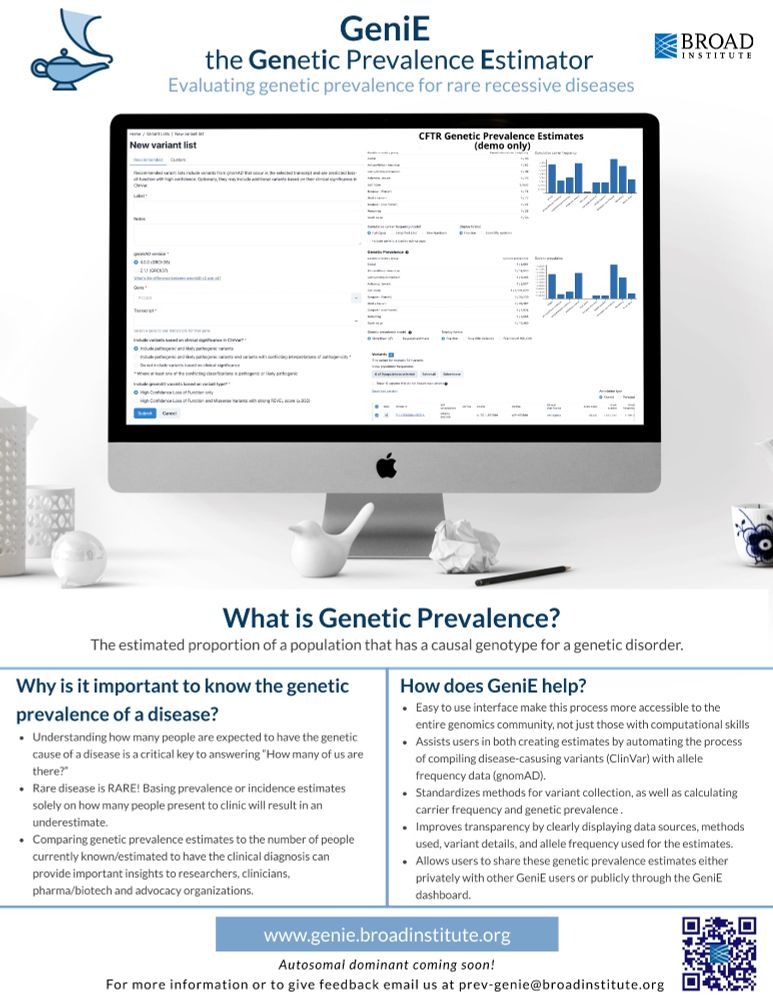
This tool allows users to estimate the genetic prevalence of autosomal recessive diseases using #gnomAD allele frequency data & classifications from #ClinVar
Blog post: broad.io/genie_blog
1) Joint AN across all called sites in exomes and genomes
2) A flag indicating when exomes and genomes frequencies are highly discordant
Learn more at broad.io/gnomad_v4-1
1) Joint AN across all called sites in exomes and genomes
2) A flag indicating when exomes and genomes frequencies are highly discordant
Learn more at broad.io/gnomad_v4-1