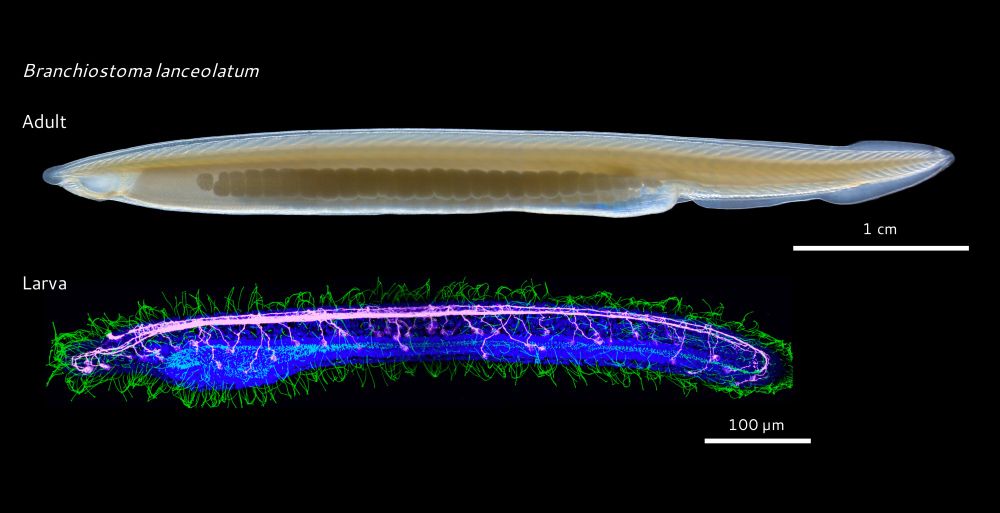
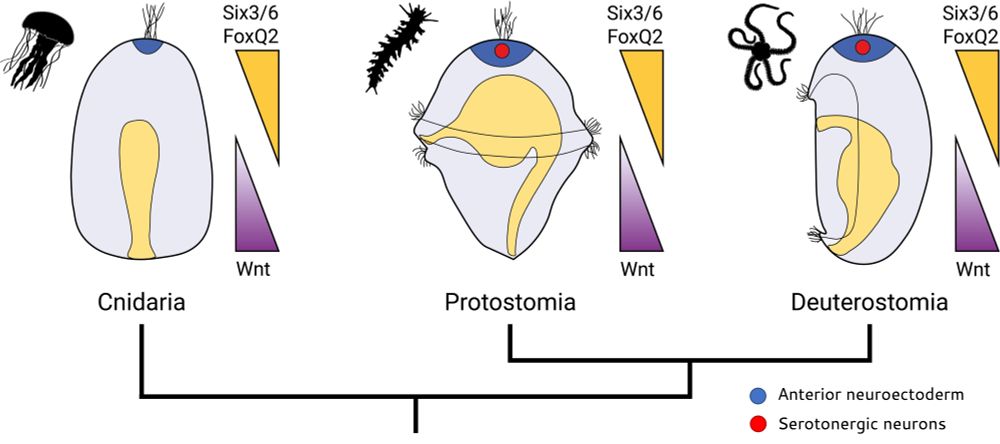
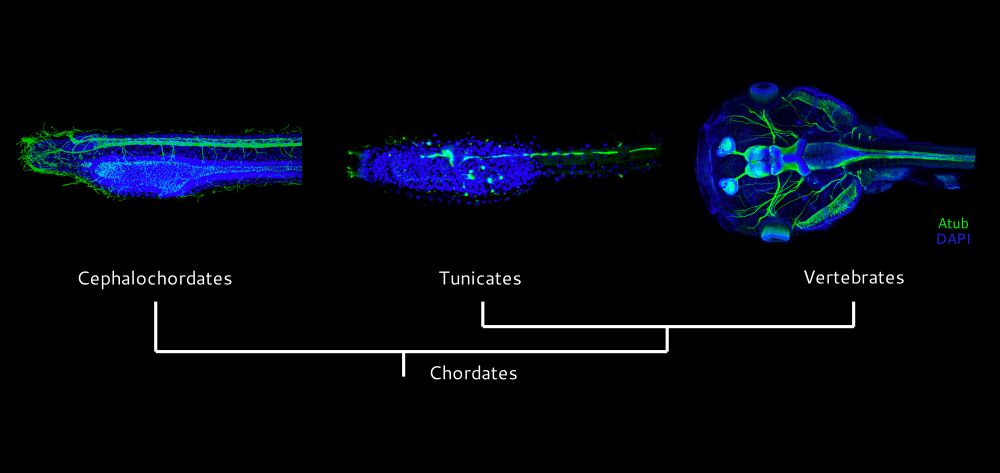
On this day we celebrate the efforts and victory of the Italian resistance movement, and the liberation of Italy from fascism and from Nazi occupation! 🎉

On this day we celebrate the efforts and victory of the Italian resistance movement, and the liberation of Italy from fascism and from Nazi occupation! 🎉