https://scholar.google.cl/citations?user=PTLeXysAAAAJ&hl=en
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
1/2
www.biorxiv.org/content/10.1...
1/2
www.encodeproject.org/single-cell/...
www.encodeproject.org/single-cell/...
doi.org/10.1038/s415...
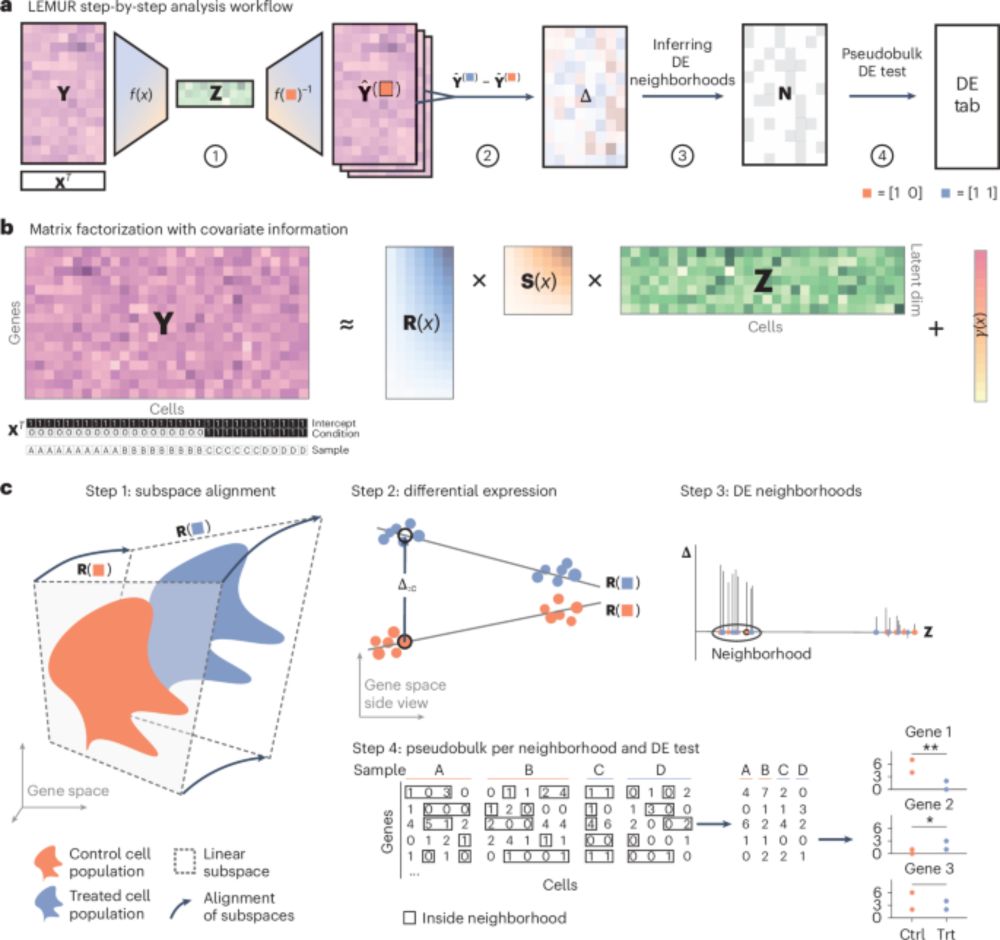
doi.org/10.1038/s415...
The largest single-cell RNA seq dataset of mouse brain aging reveals incredible insights and could pave the way for future therapies to slow or manage the impacts of the aging process. 🧵 #studyBRAIN

The largest single-cell RNA seq dataset of mouse brain aging reveals incredible insights and could pave the way for future therapies to slow or manage the impacts of the aging process. 🧵 #studyBRAIN
www.cell.com/cell/fullte...
www.cell.com/cell/fullte...
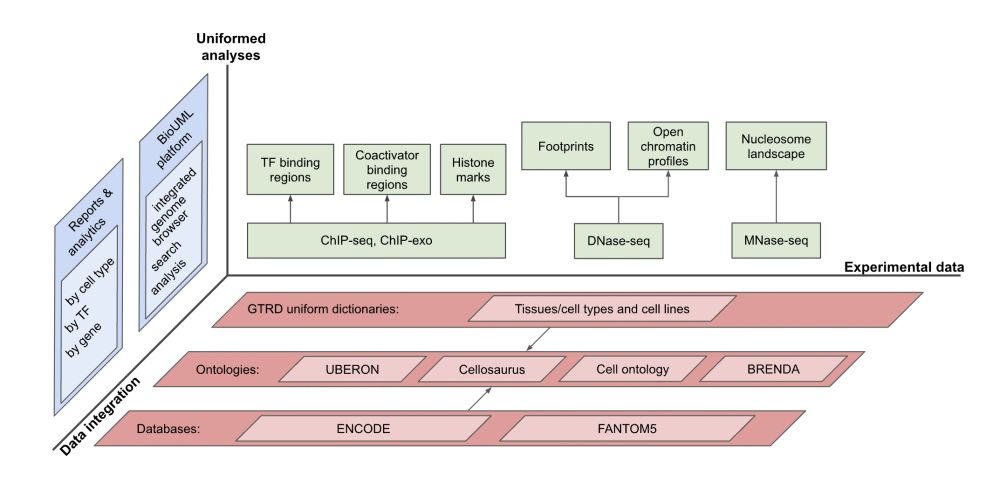
1. Is not correlated with the amount of funding
2. Progresses slowly, usually taking many years
3. Leads to more questions than it answers
4. Advances when we disengage & in improvisational discussions
5. Is too important a thing to be done in a non-playful way
1. Is not correlated with the amount of funding
2. Progresses slowly, usually taking many years
3. Leads to more questions than it answers
4. Advances when we disengage & in improvisational discussions
5. Is too important a thing to be done in a non-playful way
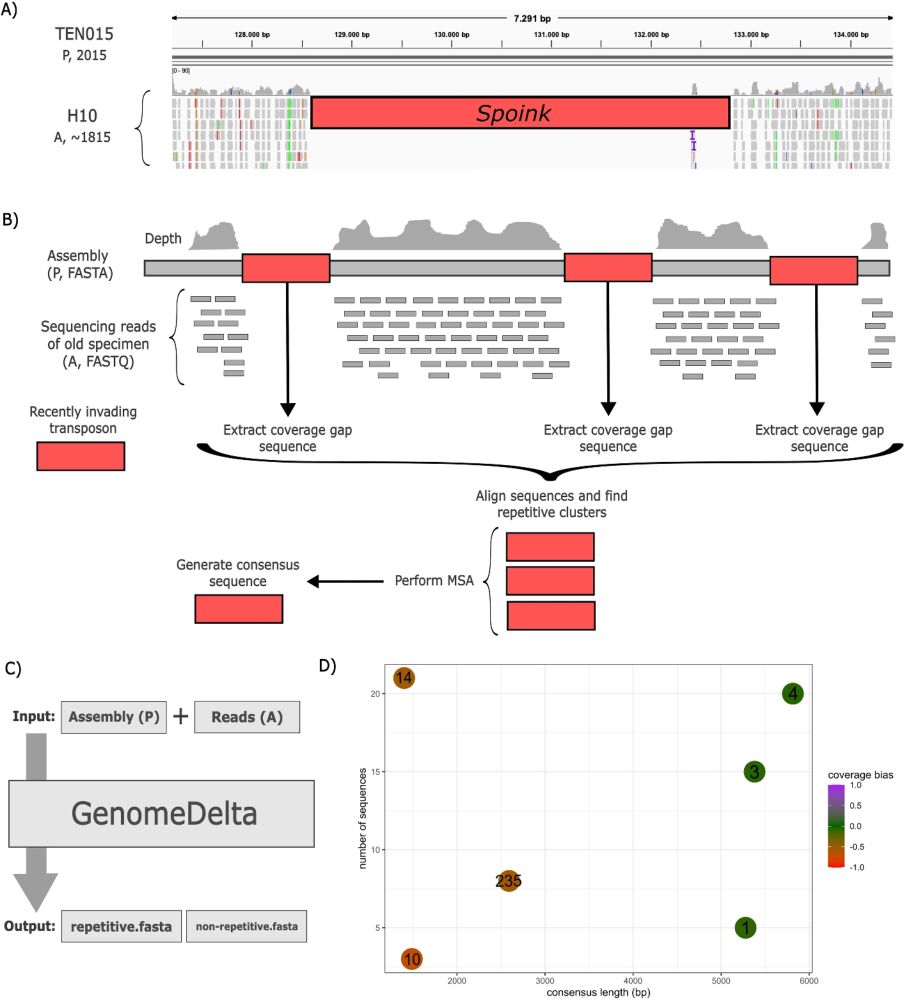
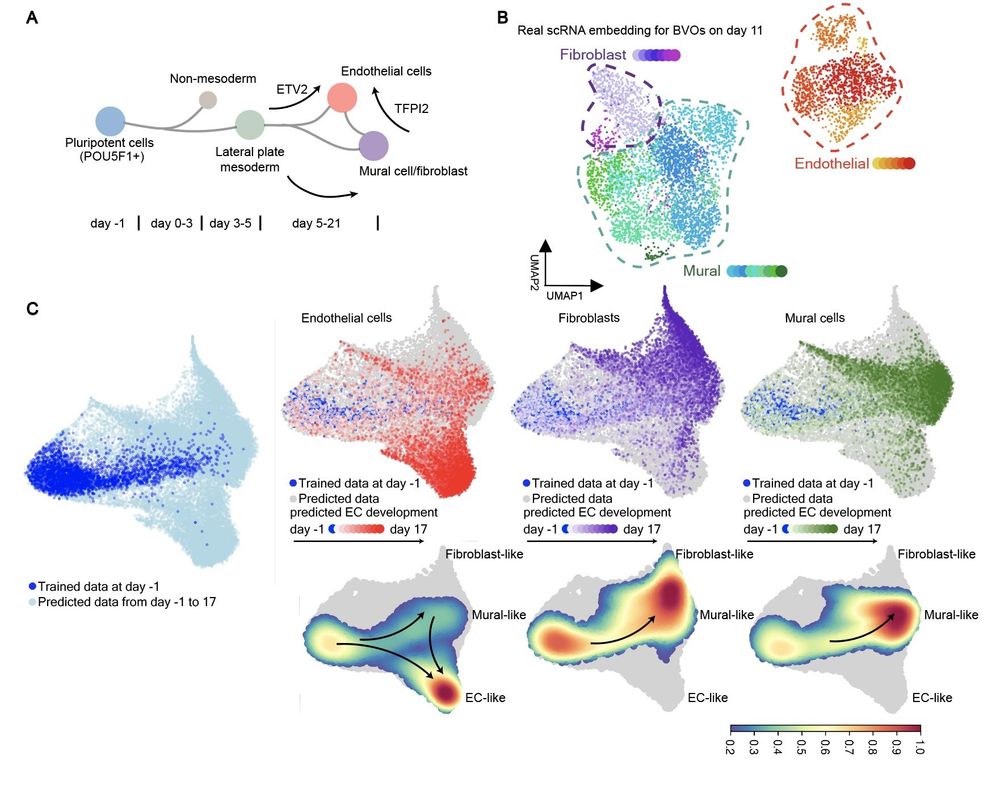


1/
www.nature.com/articles/s41...
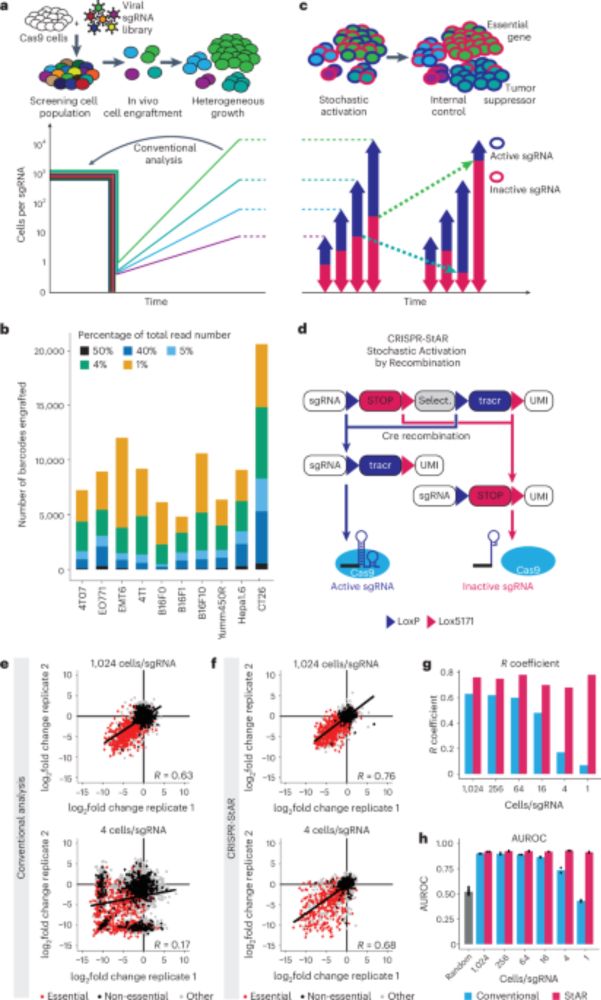
1/
www.nature.com/articles/s41...