Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
Our bottom line stayed: never use leave-one-out cross-validation as it has inherent train-test leakage. Consider our Rebalanced version instead!
We now also account for regression and nested cross-validation, with more extensive benchmarking.
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
Thread explaining the key points below.
journals.asm.org/doi/10.1128/...
Microbiome data is very variable, with substantial study- and batch-effects. DEBIAS-M corrects these, enabling robust and generalizable analyses.
A quick thread:
www.nature.com/articles/s41...
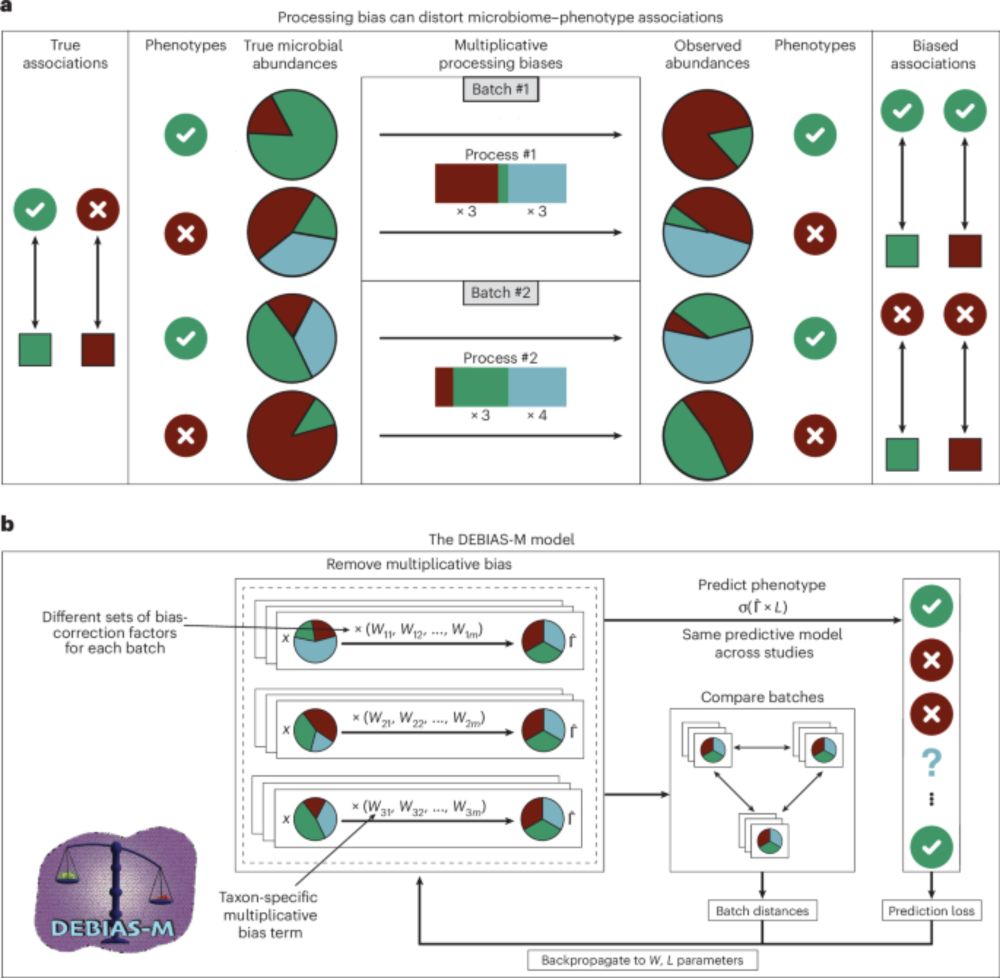
Microbiome data is very variable, with substantial study- and batch-effects. DEBIAS-M corrects these, enabling robust and generalizable analyses.
A quick thread:
www.nature.com/articles/s41...

