#LiquidBiopsy #CfRNA #RNASeq #Transcriptomics #Biomarkers #Bioinformatics #PrecisionMedicine #Preprint #CancerDiagnostics #Flomics #MethodsMatter #Reproducibility #Genomics #Diagnostics
#LiquidBiopsy #CfRNA #RNASeq #Transcriptomics #Biomarkers #Bioinformatics #PrecisionMedicine #Preprint #CancerDiagnostics #Flomics #MethodsMatter #Reproducibility #Genomics #Diagnostics
@julienlag.bsky.social, @ctuni.bsky.social, @fokkerplanck.bsky.social, @asoleee.bsky.social, Pablo Monteagudo, Elena Rusu, Lluc Cabus, Lucila González, Laura Sánchez, Beatriz Nieto & Phil Sanders who made this possible!
@julienlag.bsky.social, @ctuni.bsky.social, @fokkerplanck.bsky.social, @asoleee.bsky.social, Pablo Monteagudo, Elena Rusu, Lluc Cabus, Lucila González, Laura Sánchez, Beatriz Nieto & Phil Sanders who made this possible!
We were thrilled when the analysis revealed our own in-house Flomics libraries, using a rigorous double-DNase digestion protocol, ranked as the highest-quality dataset among all whole-cfRNA-Seq methods! 🎉💪
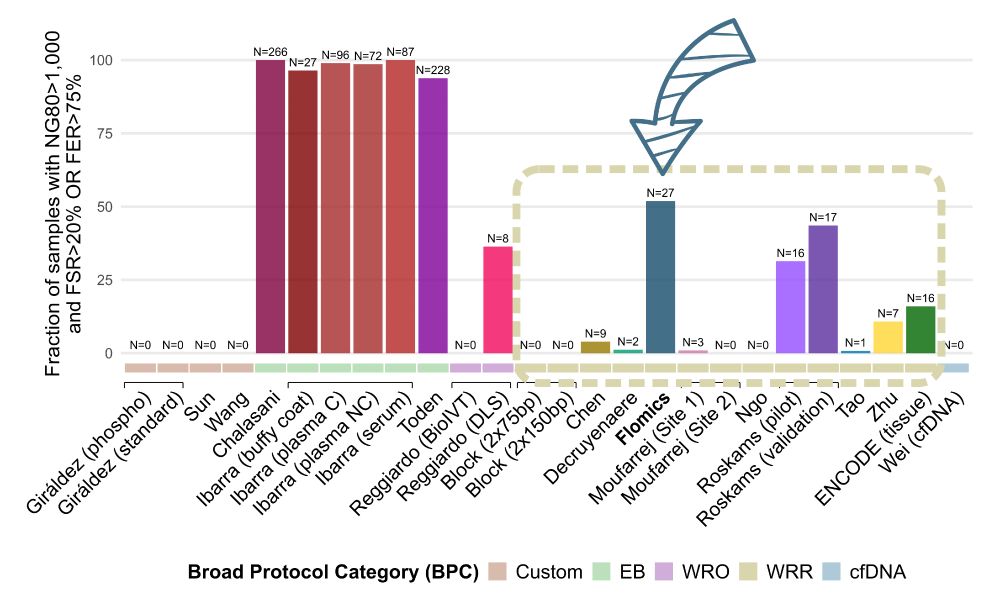
We were thrilled when the analysis revealed our own in-house Flomics libraries, using a rigorous double-DNase digestion protocol, ranked as the highest-quality dataset among all whole-cfRNA-Seq methods! 🎉💪
Our work provides a data-driven benchmark of what works, and a roadmap for more robust studies.
Our work provides a data-driven benchmark of what works, and a roadmap for more robust studies.
We highlight how technical factors are often mixed with patient phenotype, making true, generalizable biomarker discovery a huge challenge.
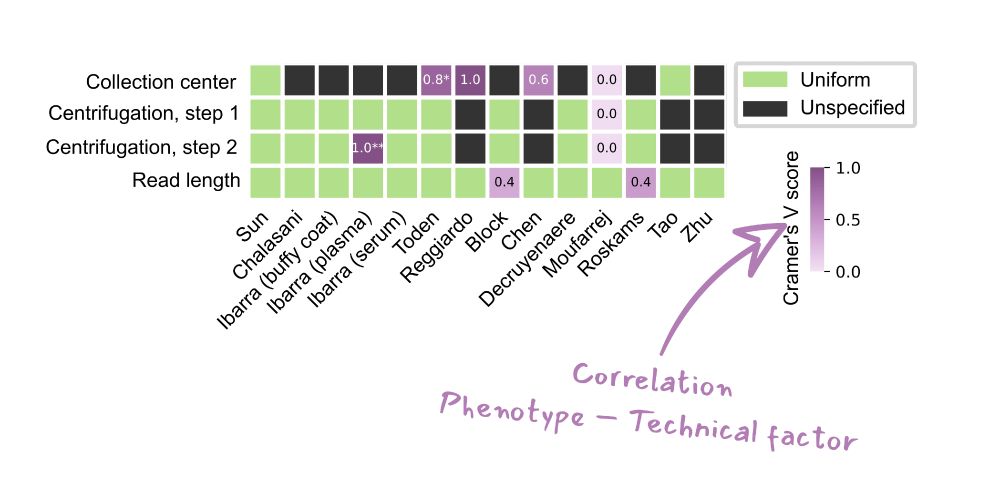
We highlight how technical factors are often mixed with patient phenotype, making true, generalizable biomarker discovery a huge challenge.
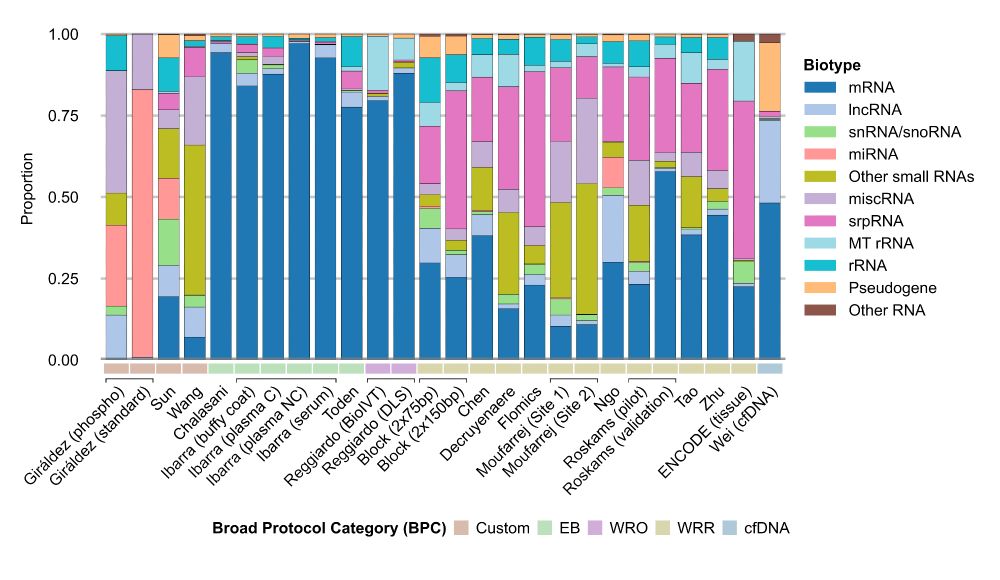
The variation we found is greater than the biological variation across 29 different human tissues. Let that sink in.
The variation we found is greater than the biological variation across 29 different human tissues. Let that sink in.
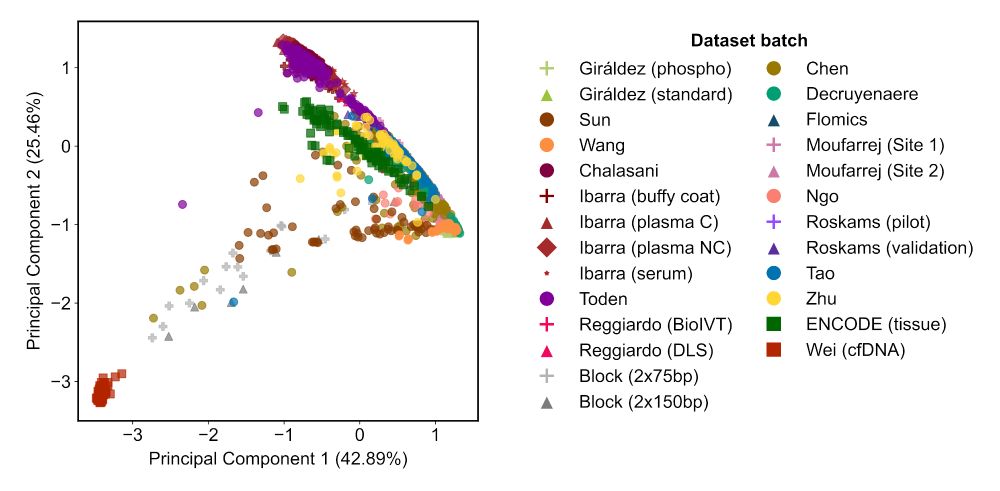
We found that inter-lab batch effects, genomic DNA contamination, and library diversity explain the main differences between samples.
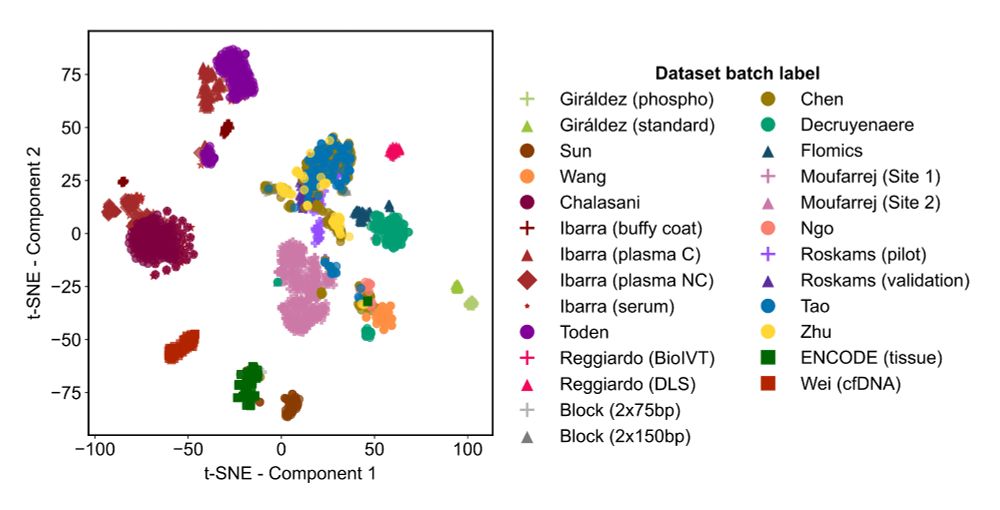
We found that inter-lab batch effects, genomic DNA contamination, and library diversity explain the main differences between samples.
Results were… illuminating.
Results were… illuminating.
We highlight how technical factors are often mixed with patient phenotype, making true, generalizable biomarker discovery a huge challenge.

We highlight how technical factors are often mixed with patient phenotype, making true, generalizable biomarker discovery a huge challenge.
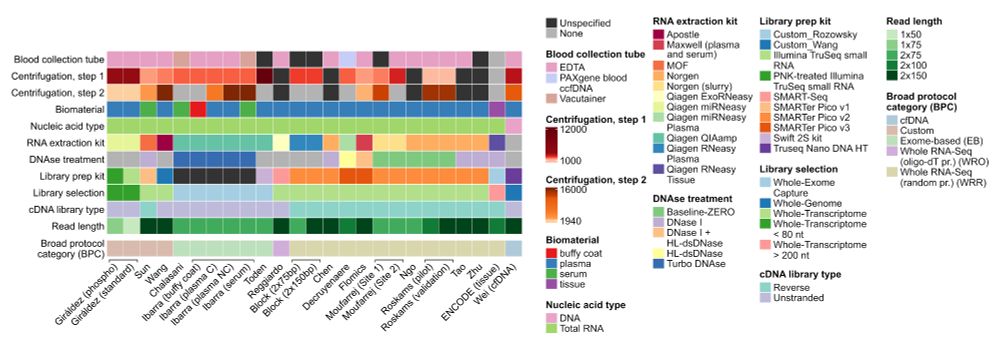
We show how different workflows give you vastly different pictures of the transcriptome.

We show how different workflows give you vastly different pictures of the transcriptome.
The variation we found is greater than the biological variation across 29 different human tissues. Let that sink in.

The variation we found is greater than the biological variation across 29 different human tissues. Let that sink in.
We found that inter-lab batch effects, genomic DNA contamination, and library diversity explain the main differences between samples.

We found that inter-lab batch effects, genomic DNA contamination, and library diversity explain the main differences between samples.

