
Felix Pförtner
@felixpfoertner.bsky.social
PhD student @enardhellmannwg.bsky.social
We combined these changes into prime-seq2. Across samples and libraries, it delivers +60% usable reads compared to prime-seq.
If you’d like to try it: we published a detailed, step-by-step protocol 🧪
👉 dx.doi.org/10.17504/pro...
🧵6/9
If you’d like to try it: we published a detailed, step-by-step protocol 🧪
👉 dx.doi.org/10.17504/pro...
🧵6/9

August 26, 2025 at 2:38 PM
We combined these changes into prime-seq2. Across samples and libraries, it delivers +60% usable reads compared to prime-seq.
If you’d like to try it: we published a detailed, step-by-step protocol 🧪
👉 dx.doi.org/10.17504/pro...
🧵6/9
If you’d like to try it: we published a detailed, step-by-step protocol 🧪
👉 dx.doi.org/10.17504/pro...
🧵6/9
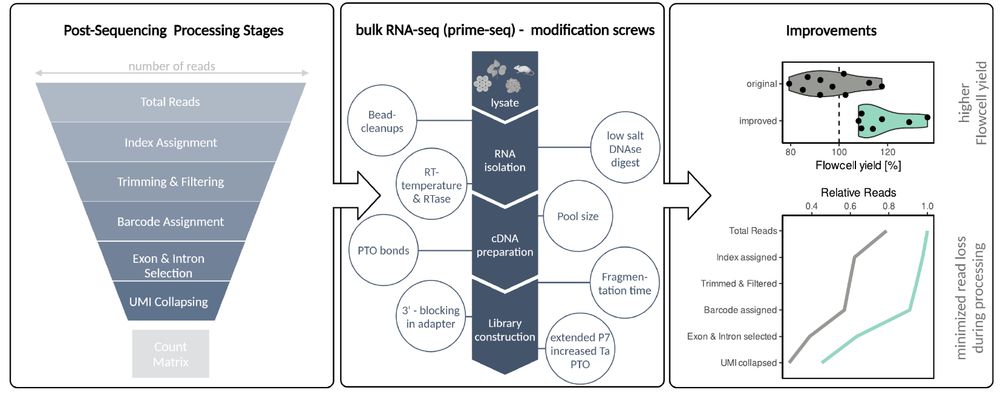
For prime-seq, the main loss points were:
• total sequencing reads
• index assignment
• intergenic reads
To improve this, we tested many alterations and ended up with:
✅ optimized DNA digest
✅ introduced PTO bonds into PCR primers
✅ redesigned ligation adapters & library PCR primers🧵5/9
• total sequencing reads
• index assignment
• intergenic reads
To improve this, we tested many alterations and ended up with:
✅ optimized DNA digest
✅ introduced PTO bonds into PCR primers
✅ redesigned ligation adapters & library PCR primers🧵5/9

August 26, 2025 at 2:38 PM
For prime-seq, the main loss points were:
• total sequencing reads
• index assignment
• intergenic reads
To improve this, we tested many alterations and ended up with:
✅ optimized DNA digest
✅ introduced PTO bonds into PCR primers
✅ redesigned ligation adapters & library PCR primers🧵5/9
• total sequencing reads
• index assignment
• intergenic reads
To improve this, we tested many alterations and ended up with:
✅ optimized DNA digest
✅ introduced PTO bonds into PCR primers
✅ redesigned ligation adapters & library PCR primers🧵5/9
This process can be nicely visualized as a funnel: at each step, reads are lost. The funnel makes it clear which stages are most responsible for read loss — and therefore where improvements could make the biggest impact.🧵4/9

August 26, 2025 at 2:38 PM
This process can be nicely visualized as a funnel: at each step, reads are lost. The funnel makes it clear which stages are most responsible for read loss — and therefore where improvements could make the biggest impact.🧵4/9
I am thrilled to finally share our new preprint on prime-seq2 🚀
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9
August 26, 2025 at 2:38 PM
I am thrilled to finally share our new preprint on prime-seq2 🚀
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9
We improved one of the most cost-efficient bulk RNAseq protocols out there to end up with +60% usable reads.
Check it out: doi.org/10.1101/2025...
a🧵1/9

