
Fabien Plisson
@fabienplisson.bsky.social
BioDesign, Machine Learning, Drug Discovery | Rosenkranz Award 2021 | Dad | Polyglot | Capybarist | plissonf.github.io
Founding ingeniebio.com
ORCID 0000-0003-224
Founding ingeniebio.com
ORCID 0000-0003-224
If that helps, we compared AF2 predicted structures of a small set of peptides against their PDB structures under different experimental methods (NMR solutions, X-ray, SSNMR, EM) as ground truth. pubs.rsc.org/en/content/a...
Benchmarking protein structure predictors to assist machine learning-guided peptide discovery
Machine learning models provide an informed and efficient strategy to create novel peptide and protein sequences with the desired profiles. Nevertheless, they are primarily trained on sequences where ...
pubs.rsc.org
June 28, 2025 at 8:25 AM
If that helps, we compared AF2 predicted structures of a small set of peptides against their PDB structures under different experimental methods (NMR solutions, X-ray, SSNMR, EM) as ground truth. pubs.rsc.org/en/content/a...
Derguini, F.; Plisson, F.; Massiot, G. 𝘗𝘳𝘦𝘱𝘢𝘳𝘢𝘵𝘪𝘰𝘯 𝘰𝘧 𝘵𝘢𝘨𝘪𝘵𝘪𝘯𝘪𝘯 𝘊 𝘢𝘯𝘥 𝘍 𝘥𝘦𝘳𝘪𝘷𝘢𝘵𝘪𝘷𝘦𝘴 𝘢𝘴 𝘢𝘯𝘵𝘪-𝘤𝘢𝘯𝘤𝘦𝘳 𝘢𝘨𝘦𝘯𝘵𝘴. FR 2941697 A1 20100806 2010.
Europe PMC europepmc.org/article/PAT/...
Google Patents
patents.google.com/patent/FR294...
Europe PMC europepmc.org/article/PAT/...
Google Patents
patents.google.com/patent/FR294...
May 5, 2025 at 1:32 AM
Derguini, F.; Plisson, F.; Massiot, G. 𝘗𝘳𝘦𝘱𝘢𝘳𝘢𝘵𝘪𝘰𝘯 𝘰𝘧 𝘵𝘢𝘨𝘪𝘵𝘪𝘯𝘪𝘯 𝘊 𝘢𝘯𝘥 𝘍 𝘥𝘦𝘳𝘪𝘷𝘢𝘵𝘪𝘷𝘦𝘴 𝘢𝘴 𝘢𝘯𝘵𝘪-𝘤𝘢𝘯𝘤𝘦𝘳 𝘢𝘨𝘦𝘯𝘵𝘴. FR 2941697 A1 20100806 2010.
Europe PMC europepmc.org/article/PAT/...
Google Patents
patents.google.com/patent/FR294...
Europe PMC europepmc.org/article/PAT/...
Google Patents
patents.google.com/patent/FR294...
As an engineer at heart, I wondered how we would scale up if we found a non-toxic, bioactive analogue. That led us to 𝗯𝗶𝗼𝗰𝗮𝘁𝗮𝗹𝘆𝘀𝗶𝘀 (think immobilised enzymes) - screening lipases and esterases to reach, in 1 step, a key intermediate: Taginitol C. This work was patented.
May 5, 2025 at 1:32 AM
As an engineer at heart, I wondered how we would scale up if we found a non-toxic, bioactive analogue. That led us to 𝗯𝗶𝗼𝗰𝗮𝘁𝗮𝗹𝘆𝘀𝗶𝘀 (think immobilised enzymes) - screening lipases and esterases to reach, in 1 step, a key intermediate: Taginitol C. This work was patented.
Over three years, we synthesised dozens of Taginitin C analogues, eventually streamlining the synthesis from 10 to 6 steps 𝘷𝘪𝘢 judicious protection/deprotection strategies.
May 5, 2025 at 1:32 AM
Over three years, we synthesised dozens of Taginitin C analogues, eventually streamlining the synthesis from 10 to 6 steps 𝘷𝘪𝘢 judicious protection/deprotection strategies.
The molecule was notoriously sensitive to acids, bases, nucleophiles, and UV light. Resources were limited, and we were performing multi-step syntheses on just 𝟱 𝗺𝗴 𝘀𝗰𝗮𝗹𝗲𝘀, in 𝘁𝗶𝗻𝘆 𝗿𝗼𝘂𝗻𝗱-𝗯𝗼𝘁𝘁𝗼𝗺 𝗳𝗹𝗮𝘀𝗸𝘀, working in darkened fume hoods.
May 5, 2025 at 1:32 AM
The molecule was notoriously sensitive to acids, bases, nucleophiles, and UV light. Resources were limited, and we were performing multi-step syntheses on just 𝟱 𝗺𝗴 𝘀𝗰𝗮𝗹𝗲𝘀, in 𝘁𝗶𝗻𝘆 𝗿𝗼𝘂𝗻𝗱-𝗯𝗼𝘁𝘁𝗼𝗺 𝗳𝗹𝗮𝘀𝗸𝘀, working in darkened fume hoods.
Together with various teammates (MSc students) we worked on the hit-lead optimisation of Tagitinin C - a toxic germanocrolide inhibiting the ubiquitin-proteasome pathway.
May 5, 2025 at 1:32 AM
Together with various teammates (MSc students) we worked on the hit-lead optimisation of Tagitinin C - a toxic germanocrolide inhibiting the ubiquitin-proteasome pathway.
Eighteen years ago, I joined one of my first drug development R&D programs at the 𝘐𝘯𝘴𝘵𝘪𝘵𝘶𝘵 𝘥𝘦𝘴 𝘚𝘤𝘪𝘦𝘯𝘤𝘦𝘴 𝘦𝘵 𝘛𝘦𝘤𝘩𝘯𝘰𝘭𝘰𝘨𝘪𝘦𝘴 𝘥𝘶 𝘔𝘦𝘥𝘪𝘤𝘢𝘮𝘦𝘯𝘵 𝘥𝘦 𝘛𝘰𝘶𝘭𝘰𝘶𝘴𝘦 - a public-private partnership between the @cnrs.fr and Pierre Fabre Laboratories.
May 5, 2025 at 1:32 AM
Eighteen years ago, I joined one of my first drug development R&D programs at the 𝘐𝘯𝘴𝘵𝘪𝘵𝘶𝘵 𝘥𝘦𝘴 𝘚𝘤𝘪𝘦𝘯𝘤𝘦𝘴 𝘦𝘵 𝘛𝘦𝘤𝘩𝘯𝘰𝘭𝘰𝘨𝘪𝘦𝘴 𝘥𝘶 𝘔𝘦𝘥𝘪𝘤𝘢𝘮𝘦𝘯𝘵 𝘥𝘦 𝘛𝘰𝘶𝘭𝘰𝘶𝘴𝘦 - a public-private partnership between the @cnrs.fr and Pierre Fabre Laboratories.
Beilun Wang and team recently reported the development of antimicrobial peptides combining an ensemble of DL predictors with EvoGradient, modifying candidate sequences strategically from evolutionary information, improving their antimicrobial activity and selectivity.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
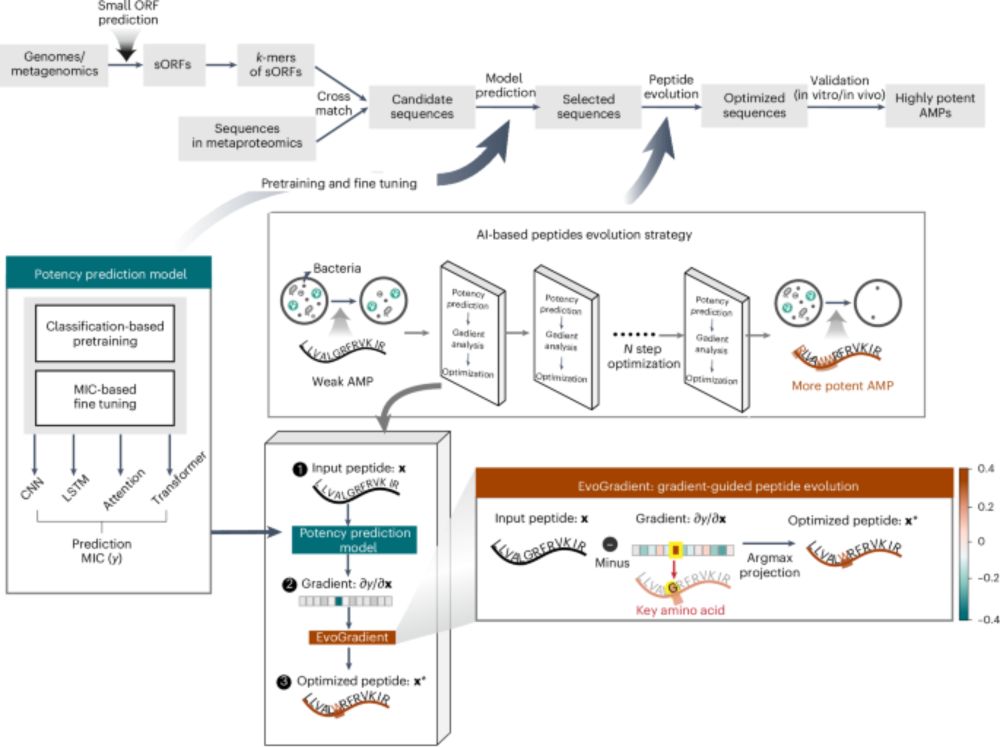
Explainable deep learning and virtual evolution identifies antimicrobial peptides with activity against multidrug-resistant human pathogens - Nature Microbiology
An AI-based learning model is applied to low-abundance human oral bacteria and identifies antimicrobial peptides with efficacy against multidrug-resistant bacterial pathogens.
www.nature.com
January 29, 2025 at 11:37 PM
Beilun Wang and team recently reported the development of antimicrobial peptides combining an ensemble of DL predictors with EvoGradient, modifying candidate sequences strategically from evolutionary information, improving their antimicrobial activity and selectivity.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
We previously used that strategy to set guidelines for discovering and designing non-hemolytic peptides:
www.nature.com/articles/s41...
www.nature.com/articles/s41...
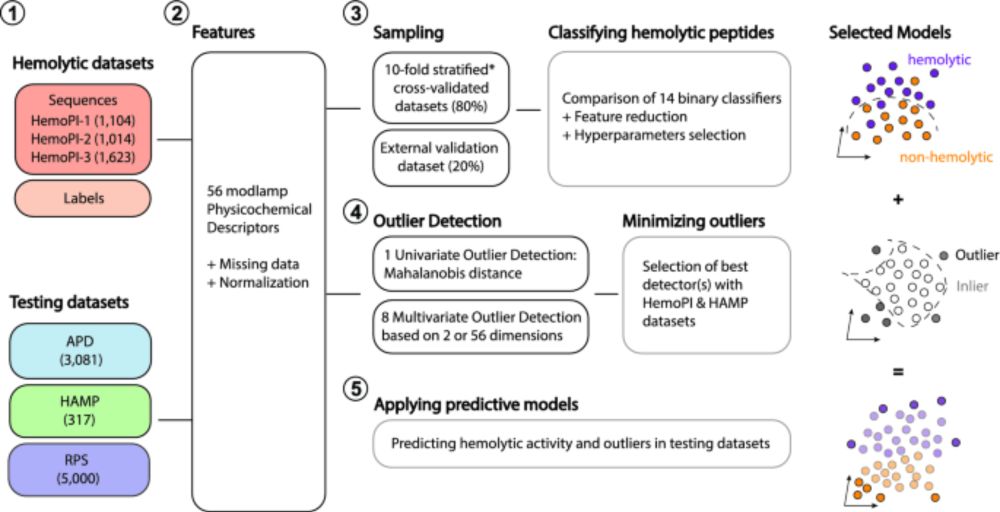
Machine learning-guided discovery and design of non-hemolytic peptides - Scientific Reports
Scientific Reports - Machine learning-guided discovery and design of non-hemolytic peptides
www.nature.com
January 29, 2025 at 11:37 PM
We previously used that strategy to set guidelines for discovering and designing non-hemolytic peptides:
www.nature.com/articles/s41...
www.nature.com/articles/s41...
The process mimics the hypothesis-driven process of a medicinal chemist, especially with Deep Mutational Scanning. It also merits explaining the results from your predictive model(s) by comparing mutants.
January 29, 2025 at 11:37 PM
The process mimics the hypothesis-driven process of a medicinal chemist, especially with Deep Mutational Scanning. It also merits explaining the results from your predictive model(s) by comparing mutants.

