@columbia prev. @harvardmed @ginkgo @yale




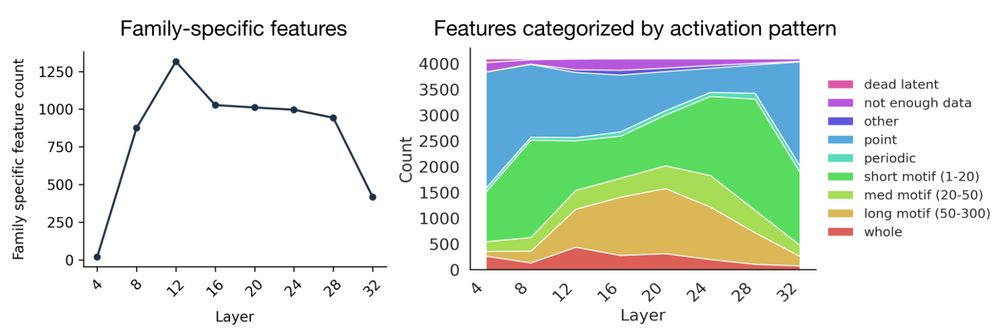
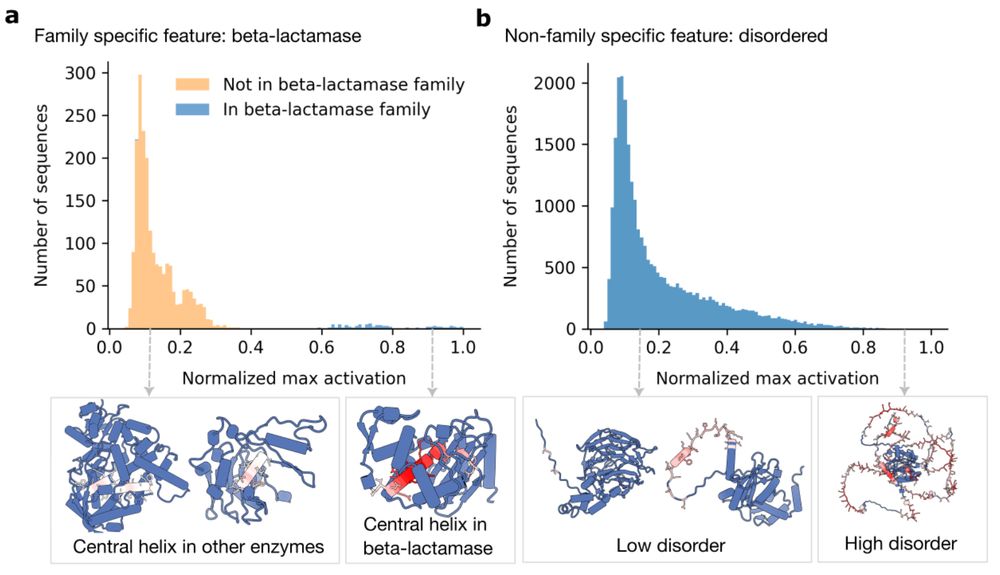
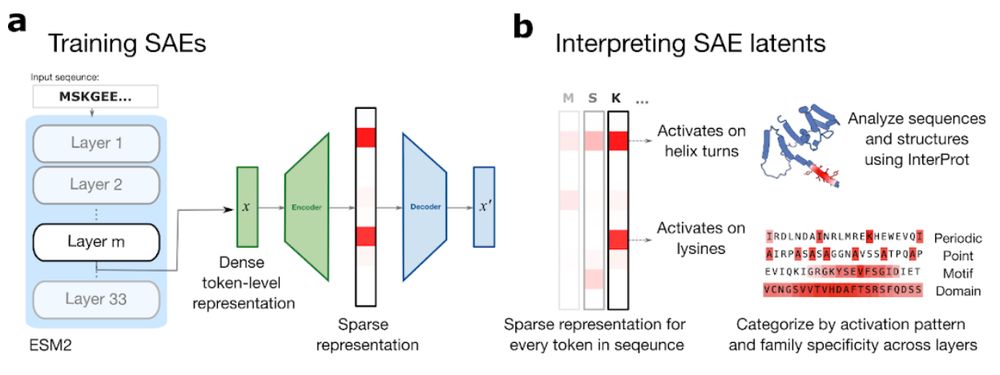
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
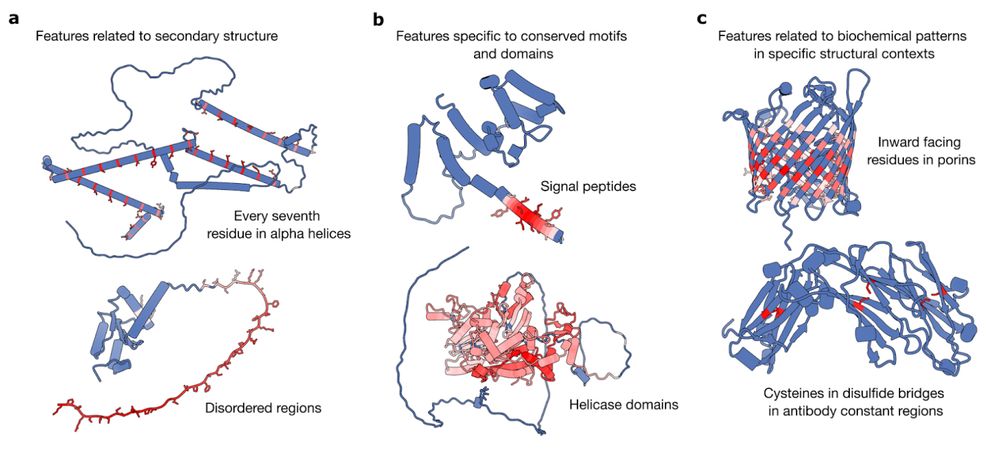
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.

