
@elihei.bsky.social and our team at @embl.org , @dkfz.bsky.social, and @mskcancercenter.bsky.social built #segger - a fast, accurate cell segmentation tool for spatial transcriptomics that assigns transcripts to their cell origins!
doi.org/10.1101/2025...

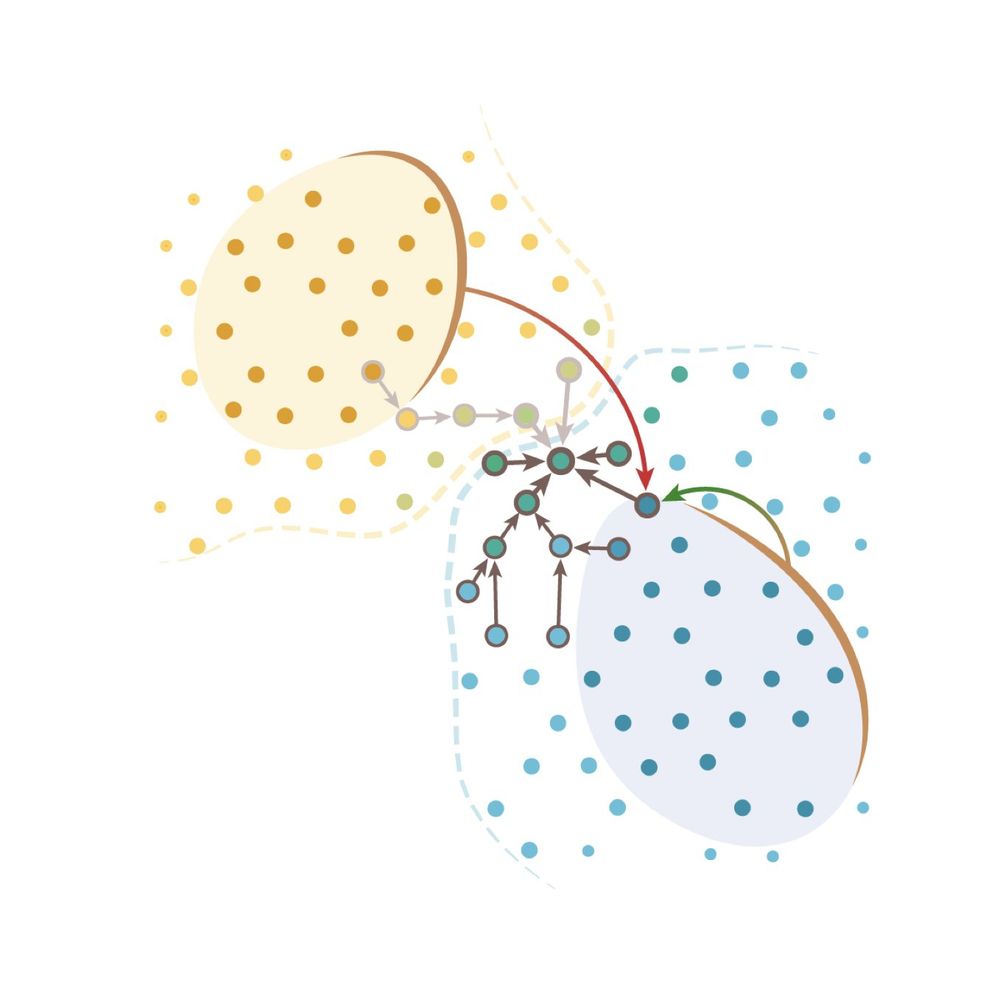
@elihei.bsky.social and our team at @embl.org , @dkfz.bsky.social, and @mskcancercenter.bsky.social built #segger - a fast, accurate cell segmentation tool for spatial transcriptomics that assigns transcripts to their cell origins!
doi.org/10.1101/2025...
Elyas Heidari and colleagues from the Gerstung, Pe'er and Stegle Labs released Segger, a new algorithm that uses GNN to model both transcripts and cells. More details in their preprint: www.biorxiv.org/content/10.1...

Elyas Heidari and colleagues from the Gerstung, Pe'er and Stegle Labs released Segger, a new algorithm that uses GNN to model both transcripts and cells. More details in their preprint: www.biorxiv.org/content/10.1...
For this reason, Elyas Heidari, a student in my lab and in @steglelab.bsky.social built segger.
www.biorxiv.org/content/10.1...
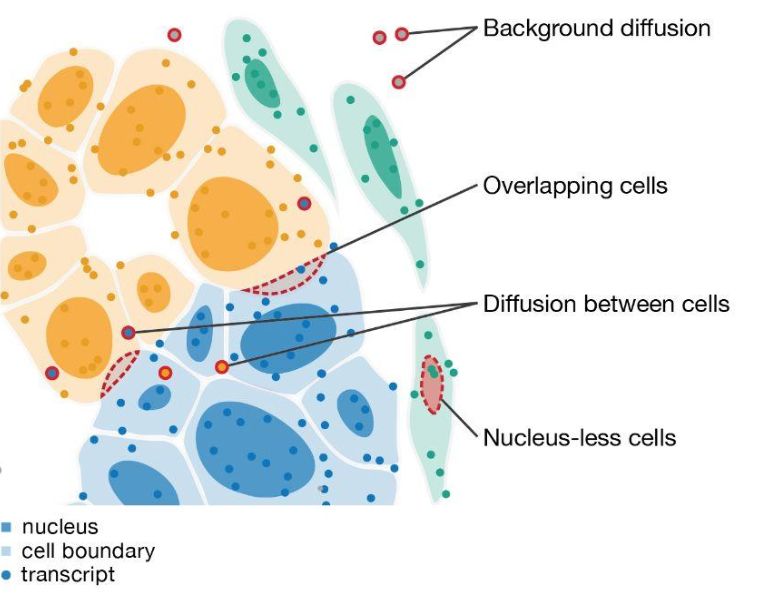
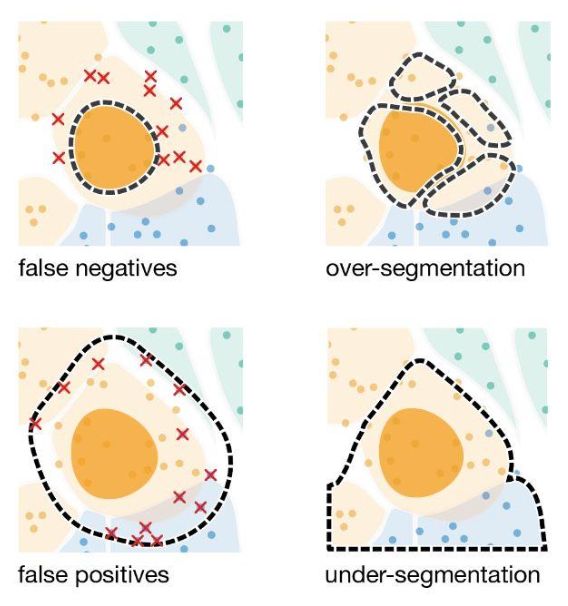
For this reason, Elyas Heidari, a student in my lab and in @steglelab.bsky.social built segger.
www.biorxiv.org/content/10.1...

