https://bits.csb.pitt.edu/






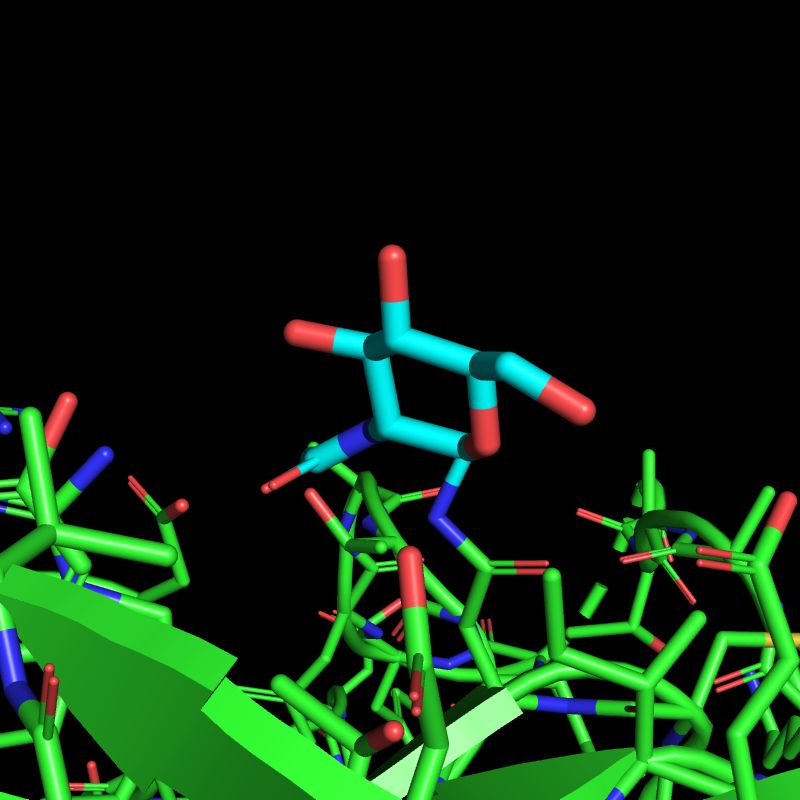















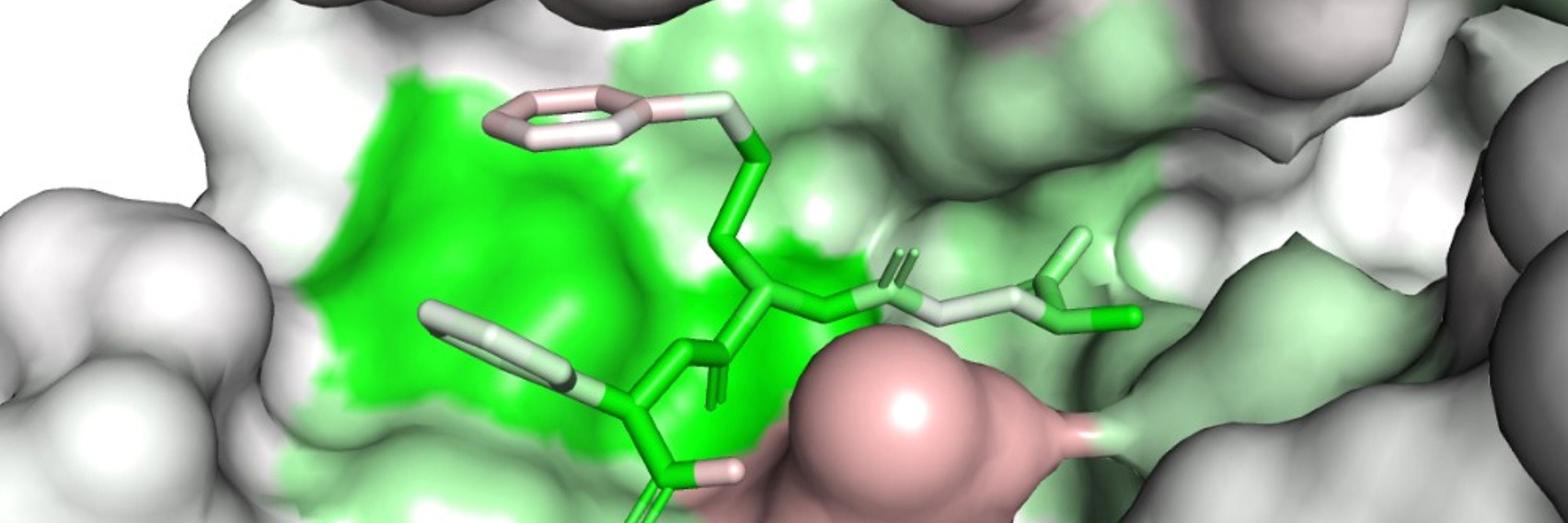
x.com/ian_dunn_/st...

x.com/ian_dunn_/st...

