Opinions my own
https://linktr.ee/ddelalamo



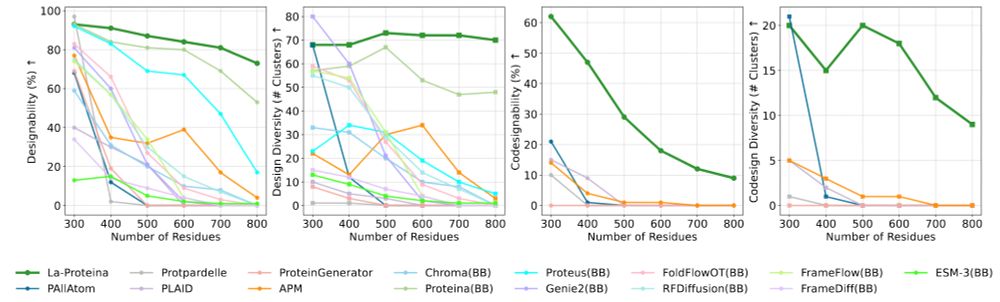
openreview.net/pdf/dd8aa97e...

openreview.net/pdf/dd8aa97e...






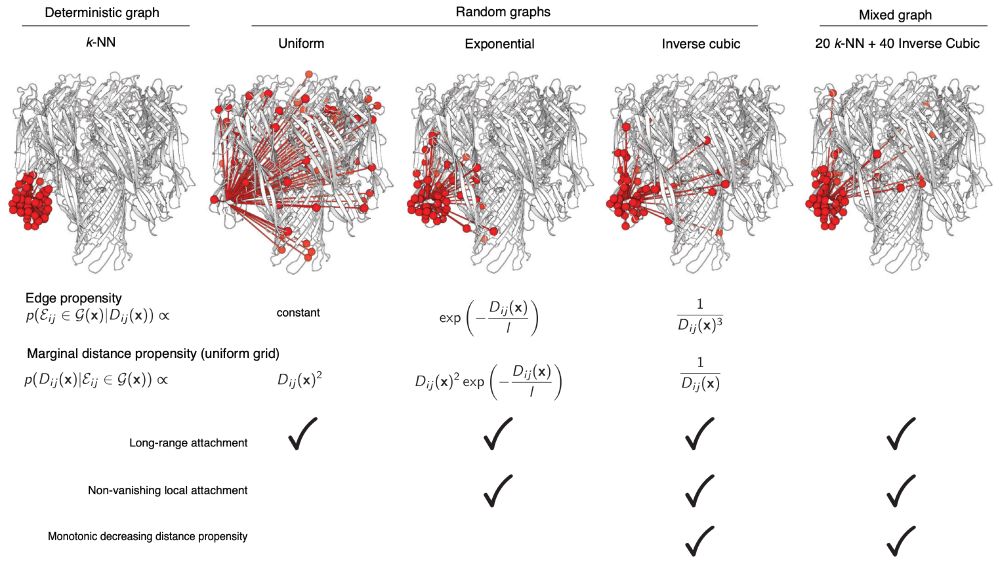
opig.stats.ox.ac.uk/webapps/sabd...

opig.stats.ox.ac.uk/webapps/sabd...