
Dani Stevens, Ph.D.
@danimstevens.bsky.social
Plant Immunologist x Machine Learning, occasional biochemist | 🌱-🧫 interactions | UC Berkeley Postdoc | 2 x USDA NIFA Fellow
100% agree. Especially with how much money is spent a year toward pub fees (10-30K+) that could go to other career opportunities. Plus, I can get high quality reviews from peers without a formal journal system. Sadly as a postdoc, I feel like my hands are tied, but I would love to help the cause.
September 16, 2025 at 6:35 AM
100% agree. Especially with how much money is spent a year toward pub fees (10-30K+) that could go to other career opportunities. Plus, I can get high quality reviews from peers without a formal journal system. Sadly as a postdoc, I feel like my hands are tied, but I would love to help the cause.
Many times, we come back to traditional journals in the end because like NIH grants, preprints don't hold the same weight/merit in trainee fellowships, tenure process, etc. It is still a challenge, though I'd like to think that if established groups move away from trad pubs, others will follow.
September 16, 2025 at 6:14 AM
Many times, we come back to traditional journals in the end because like NIH grants, preprints don't hold the same weight/merit in trainee fellowships, tenure process, etc. It is still a challenge, though I'd like to think that if established groups move away from trad pubs, others will follow.
Yes, @kseniakrasileva.bsky.social have moved to an increasingly open science model where we not only preprint early, sometimes far before journal submission, but even make datasets open before preprinting (wheat genome example: zenodo.org/records/1021...). We welcome feedback from our community.
Chromosome-level genome assembly of Triticum turgidum var 'Kronos'
This data is made available under the Toronto Agreement. All of the data listed here is available under the prepublication data sharing principle of the Toronto agreement (1). By using this data, you ...
zenodo.org
September 16, 2025 at 6:14 AM
Yes, @kseniakrasileva.bsky.social have moved to an increasingly open science model where we not only preprint early, sometimes far before journal submission, but even make datasets open before preprinting (wheat genome example: zenodo.org/records/1021...). We welcome feedback from our community.
Sweet! Those look beautiful. I wish sharing pymol/chimeraX scripts were as common as other computational work and languages.
August 18, 2025 at 3:53 AM
Sweet! Those look beautiful. I wish sharing pymol/chimeraX scripts were as common as other computational work and languages.
This is quite interesting, especially since myself and others have found the opposite (ROS, no MAPK)! If I were to $$ on it, my guess is the binding interaction in-complex is causing different PTM modifications on the kinase domain, leading to a bimodal signaling cascade. 🤔 Can't wait to read more!
July 26, 2025 at 3:39 AM
This is quite interesting, especially since myself and others have found the opposite (ROS, no MAPK)! If I were to $$ on it, my guess is the binding interaction in-complex is causing different PTM modifications on the kinase domain, leading to a bimodal signaling cascade. 🤔 Can't wait to read more!
While there's lots of work to be done, mamp-ml is a critical advancement in plant immunology for accelerating receptor-epitope characterization and engineering resistance. Mamp-ml is on Github and we implemented a version on Google Colab for easy use. Please check it out!
github.com/DanielleMSte...
github.com/DanielleMSte...
GitHub - DanielleMStevens/mamp-ml: mamp-ml: Prediction of immunogenic outcomes of LRR-PRRs and protein ligands in plants using pLM ESM-2
mamp-ml: Prediction of immunogenic outcomes of LRR-PRRs and protein ligands in plants using pLM ESM-2 - DanielleMStevens/mamp-ml
github.com
July 15, 2025 at 11:28 PM
While there's lots of work to be done, mamp-ml is a critical advancement in plant immunology for accelerating receptor-epitope characterization and engineering resistance. Mamp-ml is on Github and we implemented a version on Google Colab for easy use. Please check it out!
github.com/DanielleMSte...
github.com/DanielleMSte...
Finally, we also tested its ability to predict outcomes for convergently evolved receptors. SCORE, a csp22 receptor found in Citrus and relatives, was recently discovered by @brunongou.bsky.social. We preliminarily found mamp-ml can zero-shot predict immunogenic outcomes!

July 15, 2025 at 11:28 PM
Finally, we also tested its ability to predict outcomes for convergently evolved receptors. SCORE, a csp22 receptor found in Citrus and relatives, was recently discovered by @brunongou.bsky.social. We preliminarily found mamp-ml can zero-shot predict immunogenic outcomes!
Compared to other state-of-the-art models such as AlphaFold3, mamp-ml can predict these outcomes with higher confidence, even when a solved structure is not available. We anticipate users can now screen for immunogenic outcomes of receptor-ligand variants before spending time and $$ in the lab.

July 15, 2025 at 11:28 PM
Compared to other state-of-the-art models such as AlphaFold3, mamp-ml can predict these outcomes with higher confidence, even when a solved structure is not available. We anticipate users can now screen for immunogenic outcomes of receptor-ligand variants before spending time and $$ in the lab.
Mamp-ml harnesses the power of protein language models (ESM-2) to build a classifier model for immunogenic outcomes.
Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!
Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!

July 15, 2025 at 11:28 PM
Mamp-ml harnesses the power of protein language models (ESM-2) to build a classifier model for immunogenic outcomes.
Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!
Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!
We then generated a pipeline that combines AlphaFold2 and LRR-Annotation to precisely extract the LRR ectodomain as well as generated features to improve model training. This even includes tracking which residues interface with the ligand!

July 15, 2025 at 11:28 PM
We then generated a pipeline that combines AlphaFold2 and LRR-Annotation to precisely extract the LRR ectodomain as well as generated features to improve model training. This even includes tracking which residues interface with the ligand!
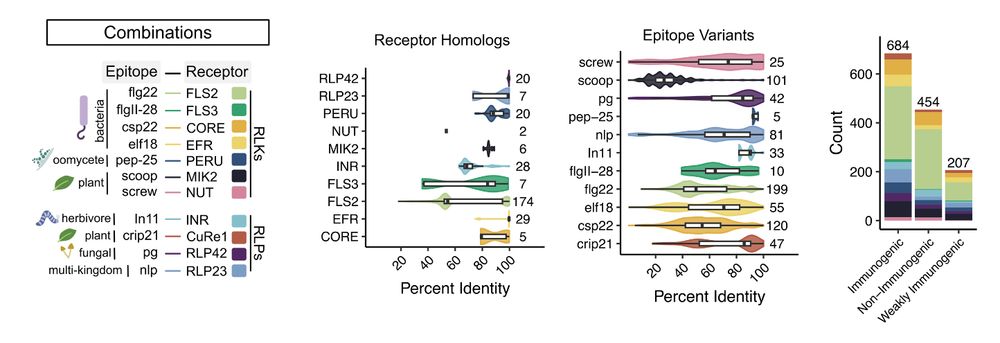
Mamp-ml was built upon two decades of foundational research. To generate the training data required, I manually pulled receptor and epitope sequences from every paper I could find, small or large. In total, we were able to capture over 1,300+ combinations across 11 receptors and 91 plant species.
July 15, 2025 at 11:28 PM
Mamp-ml was built upon two decades of foundational research. To generate the training data required, I manually pulled receptor and epitope sequences from every paper I could find, small or large. In total, we were able to capture over 1,300+ combinations across 11 receptors and 91 plant species.
It was great to catch up with so many people! Super thankful to Grey Monroe for the invite and enjoyed all the engaging questions. Can’t wait to wrap up this project and get it out there! 😁
February 4, 2025 at 5:07 AM
It was great to catch up with so many people! Super thankful to Grey Monroe for the invite and enjoyed all the engaging questions. Can’t wait to wrap up this project and get it out there! 😁
This is the one I would suggest! Was one of the first papers read as part of a phylogenetics journal club/class I took several years ago.
November 14, 2024 at 12:04 AM
This is the one I would suggest! Was one of the first papers read as part of a phylogenetics journal club/class I took several years ago.

