
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵


www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
I still wonder if there is a better solution or if someone (maybe @lionelhenry.bsky.social? 😅) would have to write a `quo_squash` function that retains the environments. :)
#Rstats
![wrap_in_tibble <- function(...){
dots <- rlang::enquos(...)
# Let's assume I can be sure that the environment is the same across dots
eval_in_mtcars(tibble::tibble(!!! dots), env = rlang::quo_get_env(dots[[1]]))
}
eval_in_mtcars <- function(expr, env){
quo <- rlang::enquo(expr)
quo <- rlang::new_quosure(rlang::quo_squash(quo), env)
rlang::eval_tidy(quo, data = mtcars[1:3,])
}
wrap_in_tibble(mpg * 3)
#> # A tibble: 3 × 1
#> `mpg * 3`
#> <dbl>
#> 1 63
#> 2 63
#> 3 68.4](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:pa4xdweapw6gnh42xqptt6vb/bafkreiedsb67npdkhwoxx3u5ceu5rfpbktvwe5dbuwqxpuledeqb43fhyu@jpeg)
I still wonder if there is a better solution or if someone (maybe @lionelhenry.bsky.social? 😅) would have to write a `quo_squash` function that retains the environments. :)
#Rstats

(Context in stackoverflow.com/questions/79...)
#Rstats #rlang
![wrap_in_df <- function(...){
dots <- rlang::enquos(...)
eval_in_mtcars(data.frame(!!! dots))
}
wrap_in_tibble <- function(...){
dots <- rlang::enquos(...)
eval_in_mtcars(tibble::tibble(!!! dots))
}
eval_in_mtcars <- function(expr){
quo <- rlang::enquo(expr)
rlang::eval_tidy(quo, data = mtcars[1:3,])
}
wrap_in_df(mpg * 2, cyl + 3)
#> X.mpg...2 X.cyl...3
#> 1 42.0 9
#> 2 42.0 9
#> 3 45.6 7
wrap_in_tibble(mpg * 2, cyl + 3)
#> Error: object 'mpg' not found](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:pa4xdweapw6gnh42xqptt6vb/bafkreihxqsxhceomx7l4lv6g5zk5ocw4vy7mlpyxf7fg5qssa5gnvjbe4q@jpeg)
(Context in stackoverflow.com/questions/79...)
#Rstats #rlang
It produces figures with consistent font size, Latex labels, and millimeter-perfect layouting. It's an alternative to patchwork with less elegant syntax but much more flexibility.
#rstats
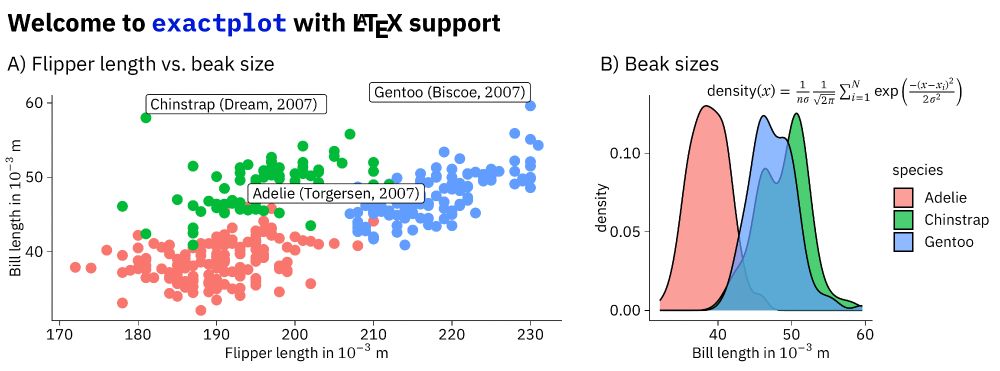
It produces figures with consistent font size, Latex labels, and millimeter-perfect layouting. It's an alternative to patchwork with less elegant syntax but much more flexibility.
#rstats
My trick is to use `tikzDevice` and do all the rendering with Luatex. I can position every panel and label with millimeter precision and still modify the output with Illustrator.

My trick is to use `tikzDevice` and do all the rendering with Luatex. I can position every panel and label with millimeter precision and still modify the output with Illustrator.
- a treatment vs. ctrl dataset from glioblastoma,
- Zebrafish developmental time course, and
- a spatially resolved Alzheimer dataset
where we find intriguing DE patterns.



- a treatment vs. ctrl dataset from glioblastoma,
- Zebrafish developmental time course, and
- a spatially resolved Alzheimer dataset
where we find intriguing DE patterns.
- close together in cell-type space, and
- have maximal differential expression.
This aggregation improves DE detection as we can optimally adjust our "cluster" resolution for each gene.

- close together in cell-type space, and
- have maximal differential expression.
This aggregation improves DE detection as we can optimally adjust our "cluster" resolution for each gene.
On top, this means you don't need to worry about abundance changes within cell types.

On top, this means you don't need to worry about abundance changes within cell types.
Inspired by generalized linear models (GLMs), I implement this as solving a regression on subspaces.

Inspired by generalized linear models (GLMs), I implement this as solving a regression on subspaces.
R package: github.com/const-ae/lemur
Python package: pylemur.readthedocs.io/en/latest/
Code from the paper: github.com/const-ae/lem...

R package: github.com/const-ae/lemur
Python package: pylemur.readthedocs.io/en/latest/
Code from the paper: github.com/const-ae/lem...
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
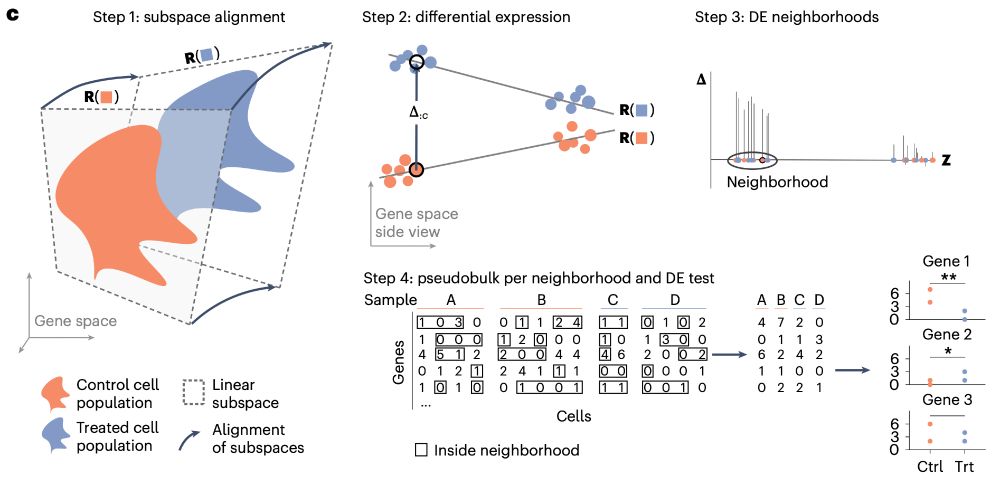
www.nature.com/articles/s41...
LEMUR is a tool to analyze multi-condition single-cell data and model differential expression as a continuous function of the cell-state space.
Some highlights⬇️
But current tools for perturbation prediction perform worse than simple linear models! We need more careful benchmarking to make progress.
www.biorxiv.org/content/10.1...
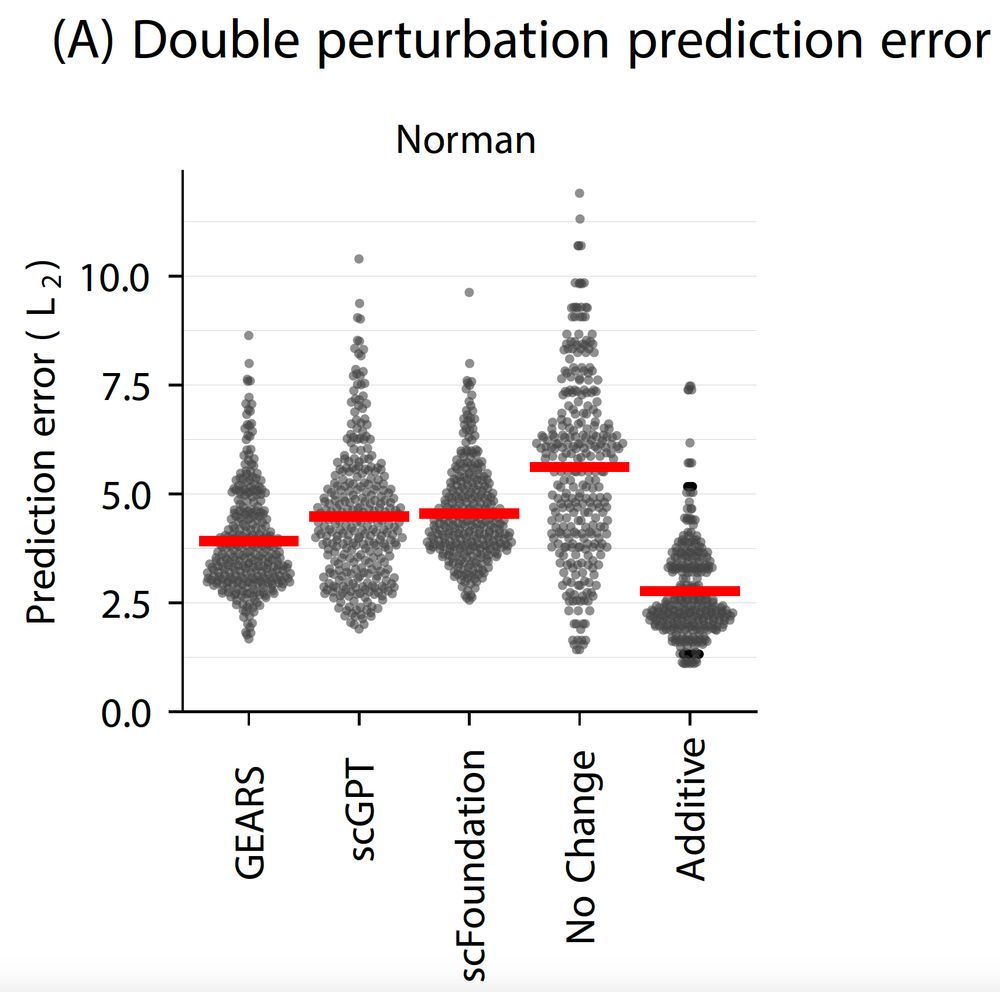
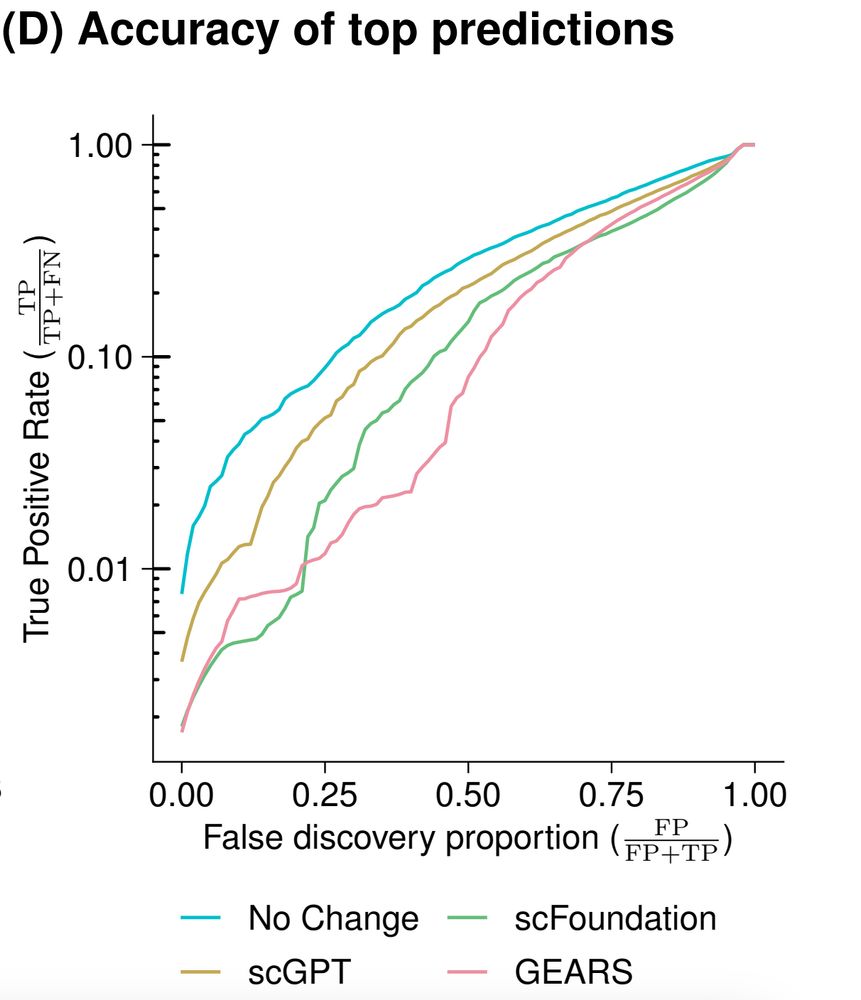
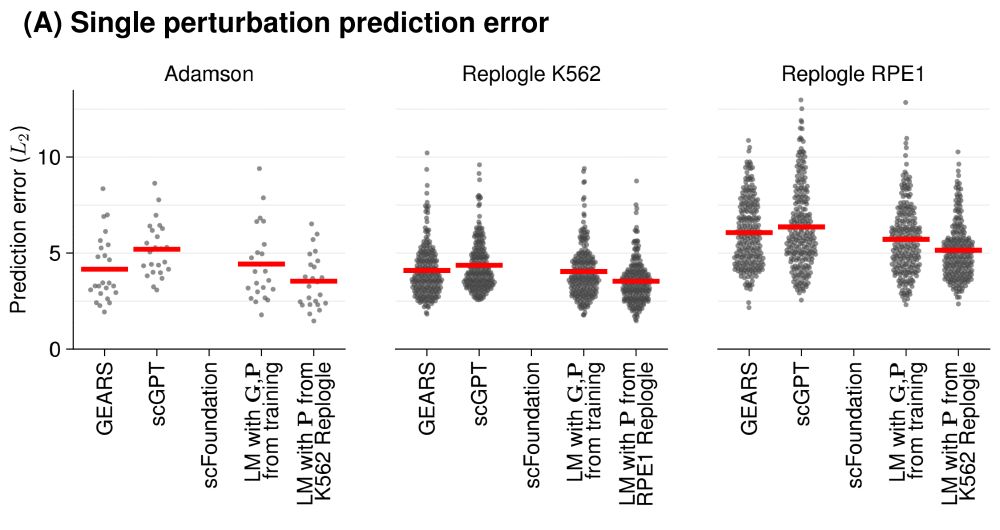
But current tools for perturbation prediction perform worse than simple linear models! We need more careful benchmarking to make progress.
www.biorxiv.org/content/10.1...
So, if you are in London and want to meet for a coffee, hit me up :)
So, if you are in London and want to meet for a coffee, hit me up :)
Now you can, for example, easily distinguish groups of discrete labels without facetting!
#rstats

Now you can, for example, easily distinguish groups of discrete labels without facetting!
#rstats
The curves are defined by a set of control points and the tangents at those points. This makes each parameter directly interpretable and easy to tweak!
#rstats #ggplot2

The curves are defined by a set of control points and the tangents at those points. This makes each parameter directly interpretable and easy to tweak!
#rstats #ggplot2
Check it out at github.com/Pweidemuelle...
The package helps with design matrices that contain colinear columns. #rstats

Check it out at github.com/Pweidemuelle...
The package helps with design matrices that contain colinear columns. #rstats
* that linear methods are flexible enough to integrate multiple conditions,
* LEMUR accurately predicts gene expression counterfactuals,
* LEMUR's DE test controls the FDR!



* that linear methods are flexible enough to integrate multiple conditions,
* LEMUR accurately predicts gene expression counterfactuals,
* LEMUR's DE test controls the FDR!
1. Analysis of the interaction between latent cell state and developmental time in embryonic development.
2. Application to a spatial single-cell dataset measuring the influence of Alzheimer's plaque density on gene expression.


1. Analysis of the interaction between latent cell state and developmental time in embryonic development.
2. Application to a spatial single-cell dataset measuring the influence of Alzheimer's plaque density on gene expression.
bioconductor.org/packages/release/bioc/html/lemur.html

bioconductor.org/packages/release/bioc/html/lemur.html
www.biorxiv.org/content/10.1... 🎉🥳
LEMUR disentangles observed and latent factors of multi-condition single-cell data and finds groups of cells with consistent differential expression for each gene.
Details on the changes ⬇️


www.biorxiv.org/content/10.1... 🎉🥳
LEMUR disentangles observed and latent factors of multi-condition single-cell data and finds groups of cells with consistent differential expression for each gene.
Details on the changes ⬇️
Thanks to everyone who joined me on this journey 🎓🧪

Thanks to everyone who joined me on this journey 🎓🧪

