
From France and now in Bonnie Edinburgh
Well done Sarah !
#ISGAXV

Well done Sarah !
#ISGAXV
Sarah is leaving the Roslin and starting her group in @exeter.ac.uk, she will be missed !
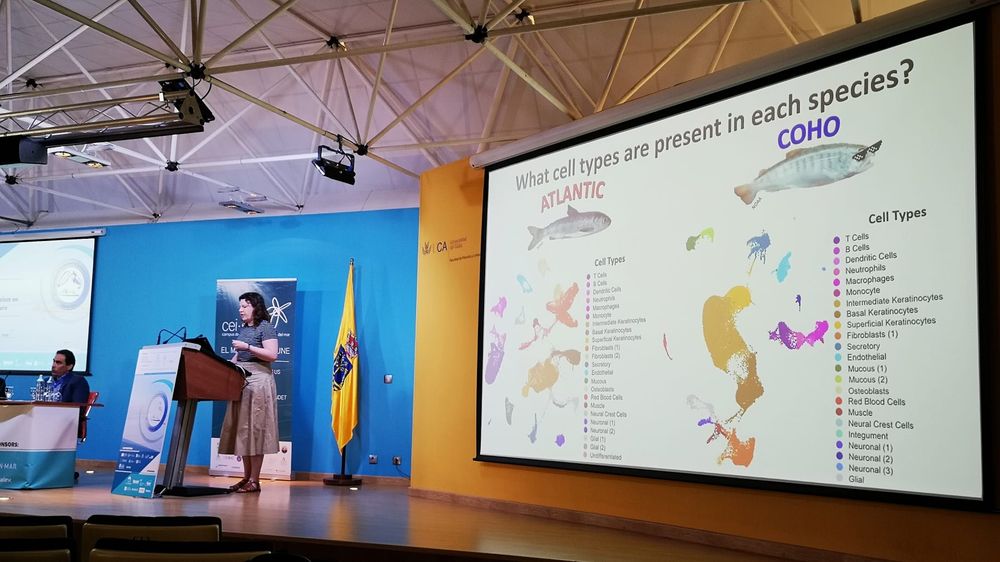
Sarah is leaving the Roslin and starting her group in @exeter.ac.uk, she will be missed !

🧪🧬🖥️

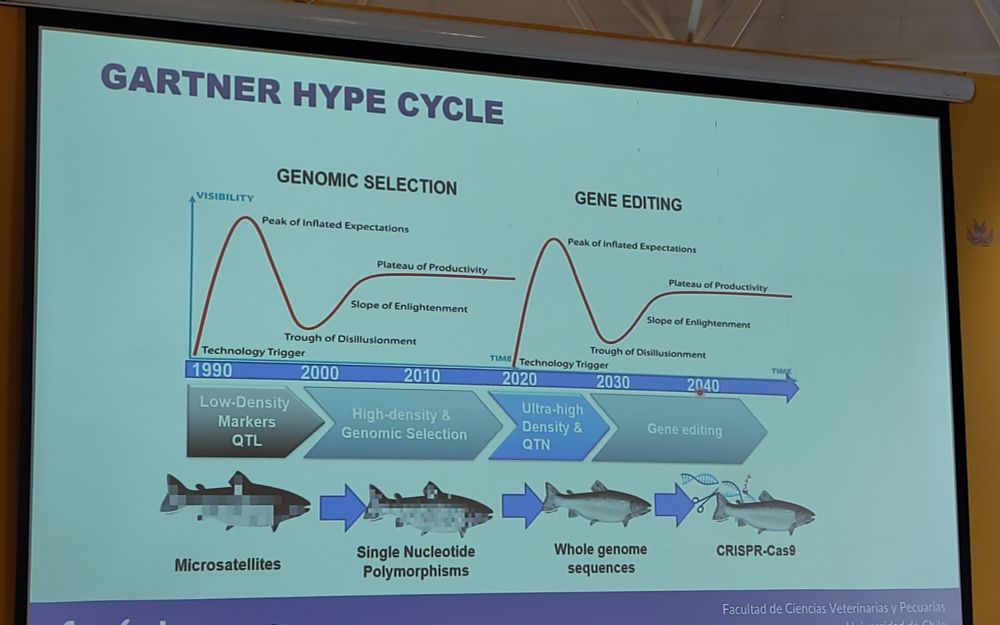
🧪🧬🖥️
There are a few posters as well to look at during the breaks and poster session 👀
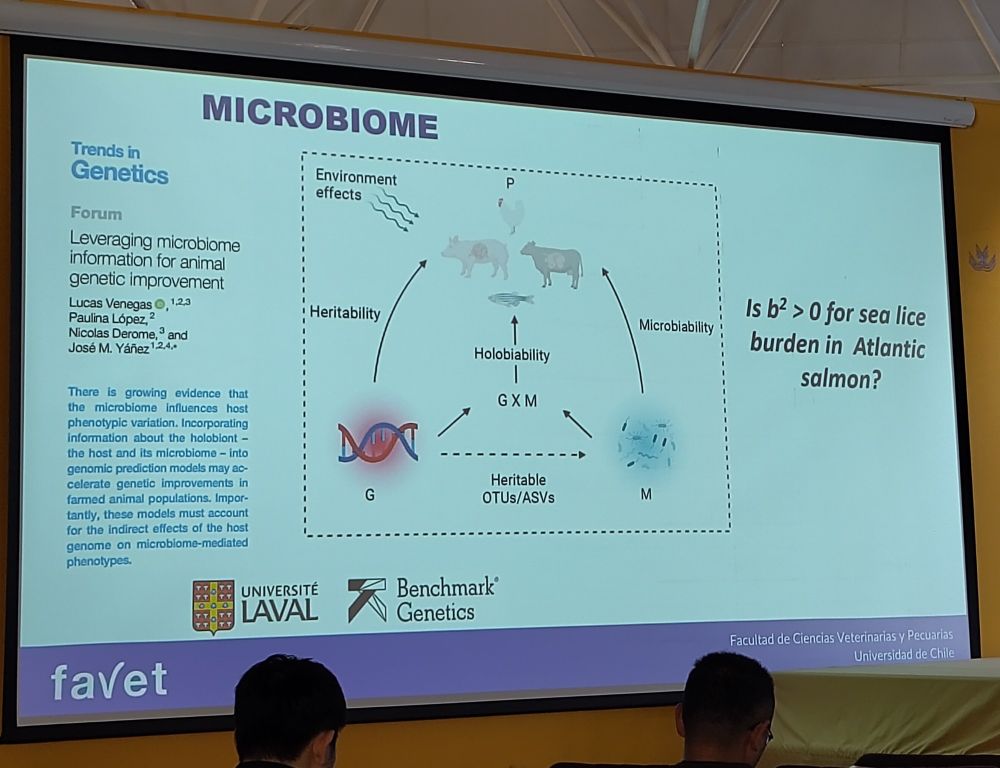
There are a few posters as well to look at during the breaks and poster session 👀
From SNP array development to implementation of genomic selection 🧬🖥️🐟
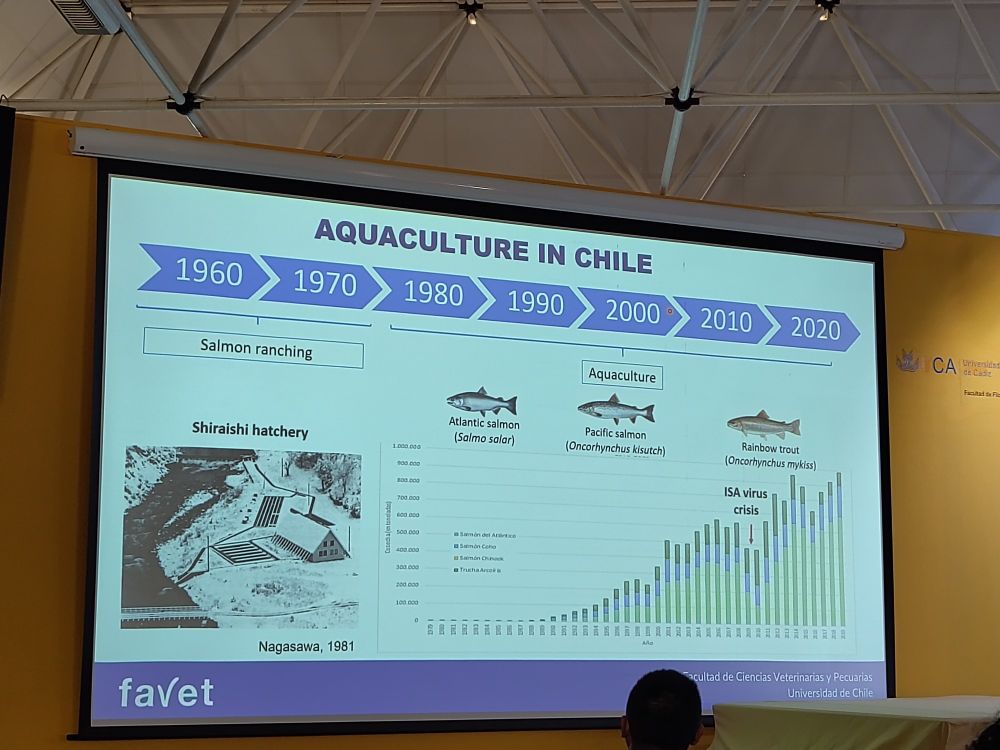
From SNP array development to implementation of genomic selection 🧬🖥️🐟

www.biorxiv.org/content/10.1...
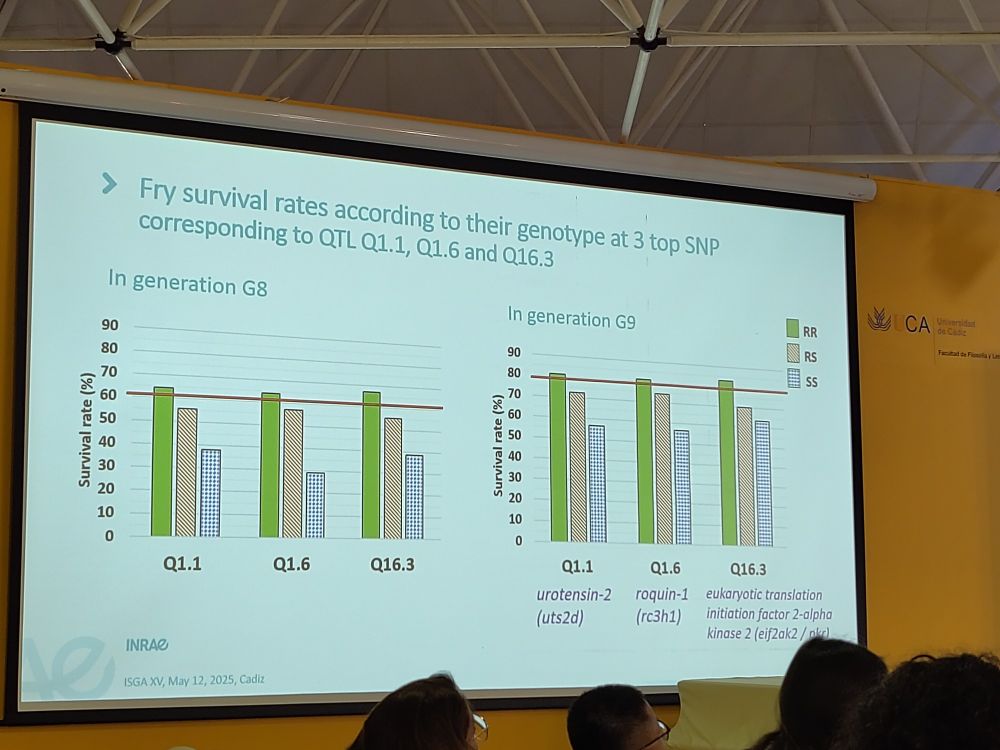
www.biorxiv.org/content/10.1...
🌨️🌧️☀️🍂

🌨️🌧️☀️🍂

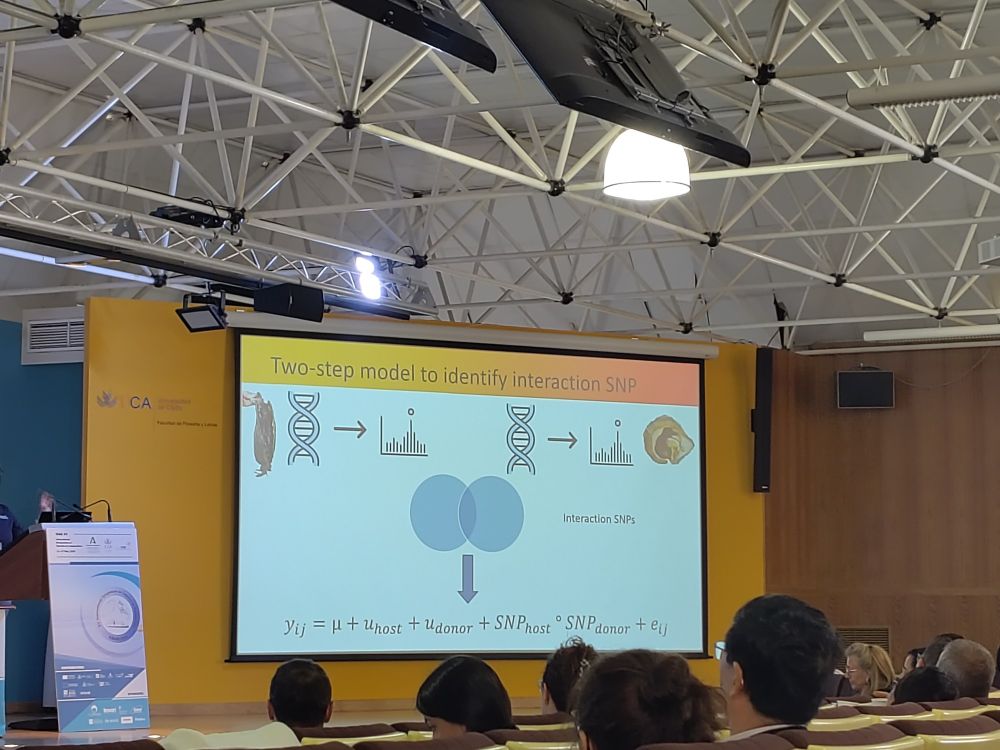
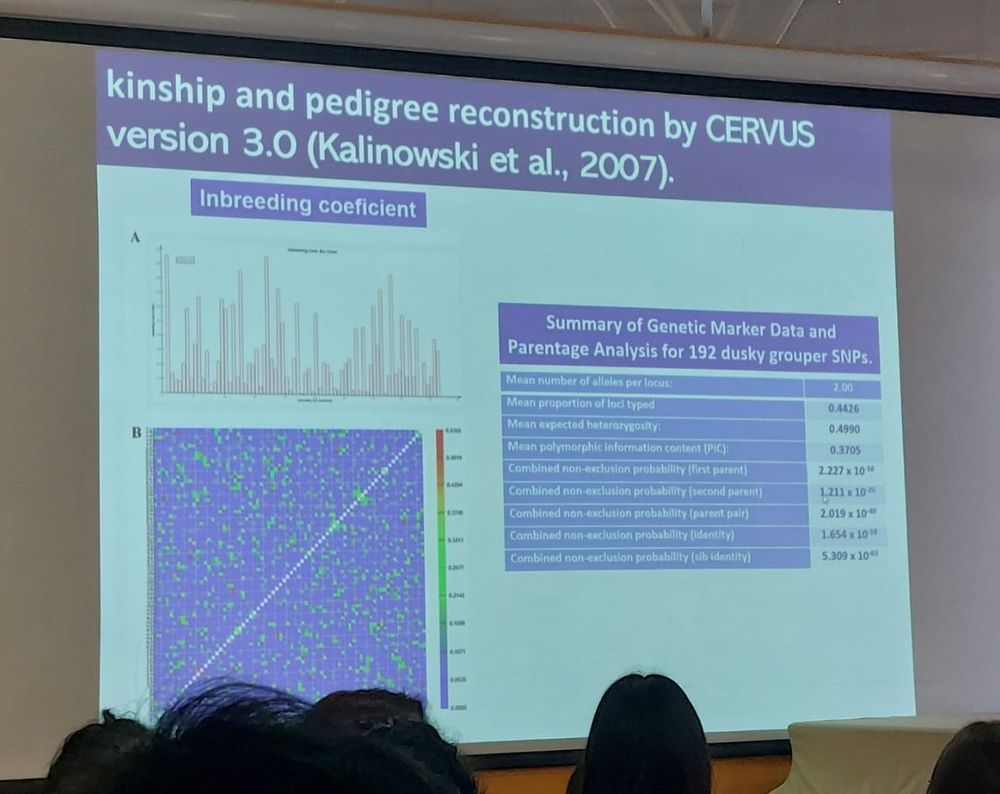
#EUAqua.org


#EUAqua.org
#ISGAXV

#ISGAXV





His group and other researchers in China worked really hard in the last 20 years to produce valuable resources 🧬🐟

His group and other researchers in China worked really hard in the last 20 years to produce valuable resources 🧬🐟


🧬🖥️

🧬🖥️

And we're in the beautiful Palazzo Bo of Padova university (where Galileo had lectures long ago, how cool is that?) @unipd.bsky.social

And we're in the beautiful Palazzo Bo of Padova university (where Galileo had lectures long ago, how cool is that?) @unipd.bsky.social

