
Group Leader at the Wallenberg Center For Molecular Medicine & SciLifeLab
https://liu.se/en/research/cantu-lab
Curious about how the genome responds to #WNT, but frankly passionate about all Science.
beyond 3D genome and open chromatin—it is the DNA Consensus sequence recognition that makes this difference!
Good old transcription factors biology.

beyond 3D genome and open chromatin—it is the DNA Consensus sequence recognition that makes this difference!
Good old transcription factors biology.



In both there is constitutively active β-catenin
How does E cells express more classical #Wnt target genes?
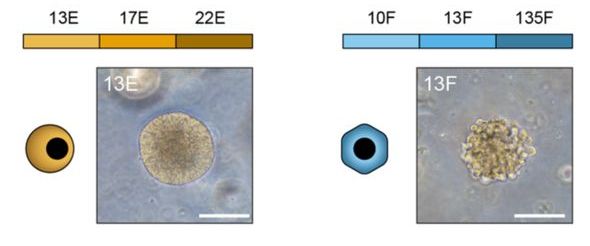

In both there is constitutively active β-catenin
How does E cells express more classical #Wnt target genes?
It shows that extracellular signals (e.g. #WNT) are acute genomic remodellers
The Genome - both sequence and 3D structure - is complex but in a matter of minutes it responds to external clues
This is a beautiful adaptation towards cell plasticity.



It shows that extracellular signals (e.g. #WNT) are acute genomic remodellers
The Genome - both sequence and 3D structure - is complex but in a matter of minutes it responds to external clues
This is a beautiful adaptation towards cell plasticity.
Yes.
The evidence?
If individual RUWs are mutated ✂️ they halve (of ~40%) the WNT>induction of ONLY that gene!

Yes.
The evidence?
If individual RUWs are mutated ✂️ they halve (of ~40%) the WNT>induction of ONLY that gene!
CTCF/Cohesin Repositioning Under Wnt (RUWs):
✅new CTCF binding sites across the genome in WNT-ON genome-wide
✅RUWs are needed for proper target gene expression
✅Yes, these include the usual suspects (e.g. AXIN2)
✅RUWs are dependent on β-catenin, which interacts with CTCF

CTCF/Cohesin Repositioning Under Wnt (RUWs):
✅new CTCF binding sites across the genome in WNT-ON genome-wide
✅RUWs are needed for proper target gene expression
✅Yes, these include the usual suspects (e.g. AXIN2)
✅RUWs are dependent on β-catenin, which interacts with CTCF
On its way to #PALS2025 in superb Göteborg!

On its way to #PALS2025 in superb Göteborg!
NPF deletion abrogates binding to #WNT nuclear complex and ability to enhance WNT transcription - BINGO!🎯
One million 💰 Q: Can we Target TBX3-NPF in human CRC?


NPF deletion abrogates binding to #WNT nuclear complex and ability to enhance WNT transcription - BINGO!🎯
One million 💰 Q: Can we Target TBX3-NPF in human CRC?

Most targets are genes known regulator of metastasis in human CRC ⬇️

Most targets are genes known regulator of metastasis in human CRC ⬇️
And impressive is the genomic co-occupancy between TBX3 and β-catenin (Fig right)


And impressive is the genomic co-occupancy between TBX3 and β-catenin (Fig right)
POP Regions:
- are found in the proximity of essential genes (KO is lethal)
- tend to be more evoutionarily conserved than other elements
- cluster aroud TSSs
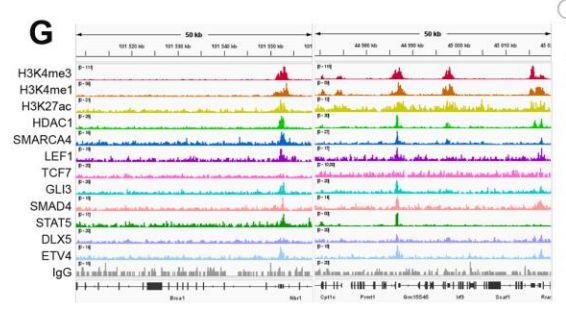


POP Regions:
- are found in the proximity of essential genes (KO is lethal)
- tend to be more evoutionarily conserved than other elements
- cluster aroud TSSs
Do you agree with us?

Do you agree with us?
Do you want to join forces and collaborate to build the future of the TranscriptionFactorBinding(TFB)-OMICS?
www.biorxiv.org/content/10.1...

Do you want to join forces and collaborate to build the future of the TranscriptionFactorBinding(TFB)-OMICS?
www.biorxiv.org/content/10.1...

