http://chaohou.netlify.app
#ProteinLM #AI4Science #ProteinDesign #MachineLearning #Bioinformatics

#ProteinLM #AI4Science #ProteinDesign #MachineLearning #Bioinformatics




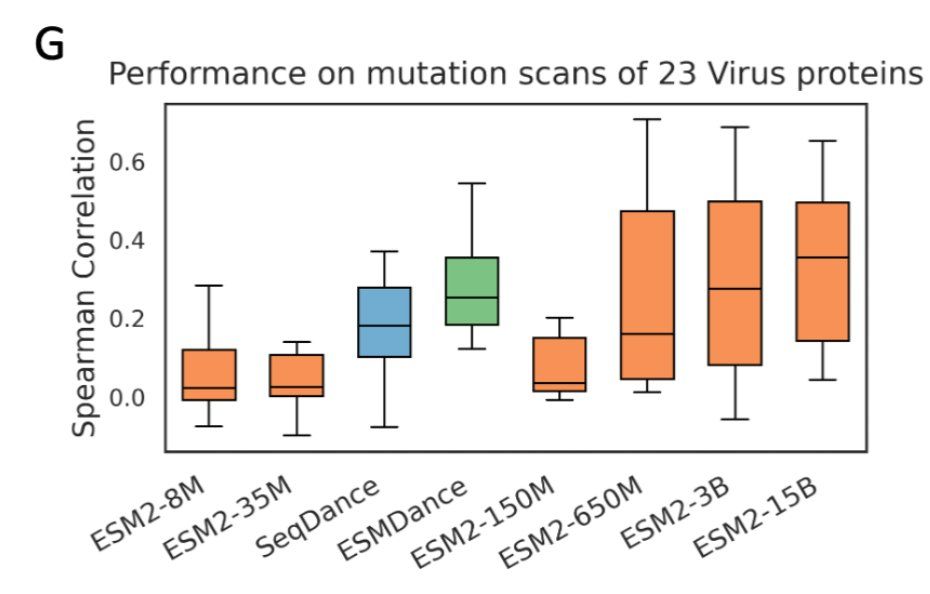
The better a pLM’s per-residue likelihoods align with MSA-based estimates, the better its performance on fitness prediction. 📈

The better a pLM’s per-residue likelihoods align with MSA-based estimates, the better its performance on fitness prediction. 📈
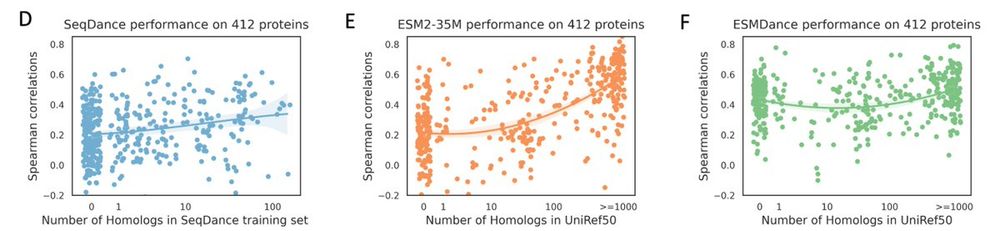
Even though pLMs are also trained to learn evolutionary information, their predicted whole sequence likelihoods show no correlation with MSA-based methods. ⚡

Even though pLMs are also trained to learn evolutionary information, their predicted whole sequence likelihoods show no correlation with MSA-based methods. ⚡
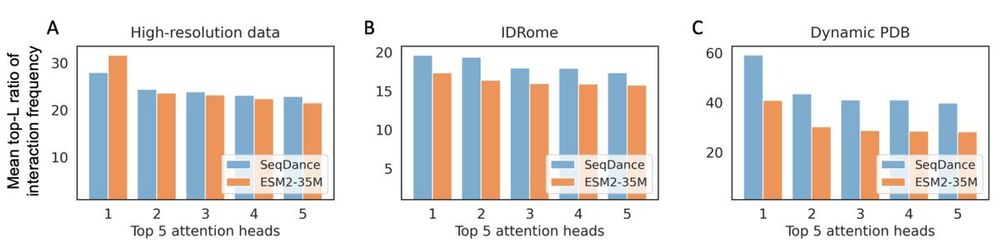
Interestingly, MSA-based models do not show this trend.

Interestingly, MSA-based models do not show this trend.
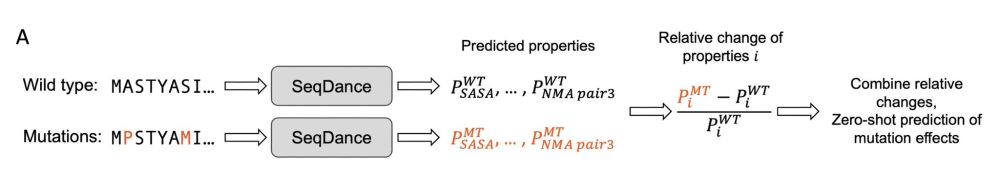
To infer mutation effects, the log-likelihood ratio (LLR) between the mutated and wild-type sequences is used. ⚖️

To infer mutation effects, the log-likelihood ratio (LLR) between the mutated and wild-type sequences is used. ⚖️