
Ben Engel
@cellarchlab.com
cellarchlab.com Univ. of Basel Biozentrum🇨🇭. Exploring molecular architecture inside cells with #CryoEM #TeamTomo. Plants and algae in a changing climate. ❄🔬 OF 🌿 4THE 🌍!
A class like no other!! From #AI structure hallucination 🤖 to #CryoEM structure reality 🔬 by @biozentrum.unibas.ch @unibas.ch undergraduate students👩🔬👩🎓 in just a few weeks!
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪

October 26, 2025 at 1:48 PM
A class like no other!! From #AI structure hallucination 🤖 to #CryoEM structure reality 🔬 by @biozentrum.unibas.ch @unibas.ch undergraduate students👩🔬👩🎓 in just a few weeks!
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
I think it has something to do with conservative lawmakers taking aim at liberal-leaning social media. See this announcement that my friend got. Did you get something similar? X is not affected by this law, and that place is swarming with misinformation and porn bots. No free speech = Hot Garbage 🔥🗑️

September 30, 2025 at 12:21 PM
I think it has something to do with conservative lawmakers taking aim at liberal-leaning social media. See this announcement that my friend got. Did you get something similar? X is not affected by this law, and that place is swarming with misinformation and porn bots. No free speech = Hot Garbage 🔥🗑️
EXCITING NEWS!! We're searching for a new professor colleague at the Biozentrum🇨🇭. Ideal candidates are exploring novel biological questions with structure, biophysics, or high-resolution imaging. 🧪 🧶🧬 🔬
The call closes Nov 3. Please get in touch if you would like more information. Come join us! 👩🔬🤓
The call closes Nov 3. Please get in touch if you would like more information. Come join us! 👩🔬🤓

September 30, 2025 at 9:52 AM
EXCITING NEWS!! We're searching for a new professor colleague at the Biozentrum🇨🇭. Ideal candidates are exploring novel biological questions with structure, biophysics, or high-resolution imaging. 🧪 🧶🧬 🔬
The call closes Nov 3. Please get in touch if you would like more information. Come join us! 👩🔬🤓
The call closes Nov 3. Please get in touch if you would like more information. Come join us! 👩🔬🤓
Love it! 🧪🤓 Ya know, thylakoids can also play this paracrystalline lattice game. In plants, they emerge from the prolamellar body during de-etiolation 🟡➡️🟢
Little piece by @wojwie.bsky.social & me:
www.nature.com/articles/s41...
Lovely review by Andrew Staehelin💚:
link.springer.com/article/10.1...
Little piece by @wojwie.bsky.social & me:
www.nature.com/articles/s41...
Lovely review by Andrew Staehelin💚:
link.springer.com/article/10.1...

September 28, 2025 at 12:11 AM
Love it! 🧪🤓 Ya know, thylakoids can also play this paracrystalline lattice game. In plants, they emerge from the prolamellar body during de-etiolation 🟡➡️🟢
Little piece by @wojwie.bsky.social & me:
www.nature.com/articles/s41...
Lovely review by Andrew Staehelin💚:
link.springer.com/article/10.1...
Little piece by @wojwie.bsky.social & me:
www.nature.com/articles/s41...
Lovely review by Andrew Staehelin💚:
link.springer.com/article/10.1...
An interesting parallel: Rod outer-segment disk membranes are highly packed with a membrane protein (rhodopsin) 👁️, similar to the stacked domains of thylakoids (PSII-LHCII) 🌱. And isolation of disk membranes may cause rhodopsin to crystallize into 2D arrays 💠
www.nature.com/articles/421... 🧪 🧶🧬🌾🔬
www.nature.com/articles/421... 🧪 🧶🧬🌾🔬


September 27, 2025 at 11:45 PM
An interesting parallel: Rod outer-segment disk membranes are highly packed with a membrane protein (rhodopsin) 👁️, similar to the stacked domains of thylakoids (PSII-LHCII) 🌱. And isolation of disk membranes may cause rhodopsin to crystallize into 2D arrays 💠
www.nature.com/articles/421... 🧪 🧶🧬🌾🔬
www.nature.com/articles/421... 🧪 🧶🧬🌾🔬
This study completes a trilogy of thylakoid papers in @elife.bsky.social, one every 5 years⏳⌛
Part 1 was my first #CryoET paper: elifesciences.org/articles/04889
Part 2 was my first paper with @wojwie.bsky.social: elifesciences.org/articles/53740
Time flies... so let's start working on Part 4? 🤓🧪
Part 1 was my first #CryoET paper: elifesciences.org/articles/04889
Part 2 was my first paper with @wojwie.bsky.social: elifesciences.org/articles/53740
Time flies... so let's start working on Part 4? 🤓🧪

September 25, 2025 at 6:59 PM
This study completes a trilogy of thylakoid papers in @elife.bsky.social, one every 5 years⏳⌛
Part 1 was my first #CryoET paper: elifesciences.org/articles/04889
Part 2 was my first paper with @wojwie.bsky.social: elifesciences.org/articles/53740
Time flies... so let's start working on Part 4? 🤓🧪
Part 1 was my first #CryoET paper: elifesciences.org/articles/04889
Part 2 was my first paper with @wojwie.bsky.social: elifesciences.org/articles/53740
Time flies... so let's start working on Part 4? 🤓🧪
So, PSII arrays seem to be an artifact of isolating thylakoid membranes. And yet… We still wonder if these beautiful (and likely more static) structures play some physiological role during stress or damage. @wojwie.bsky.social & I wrote a mini-review on this:
academic.oup.com/plcell/artic...
12/🧵
academic.oup.com/plcell/artic...
12/🧵

September 25, 2025 at 6:49 PM
So, PSII arrays seem to be an artifact of isolating thylakoid membranes. And yet… We still wonder if these beautiful (and likely more static) structures play some physiological role during stress or damage. @wojwie.bsky.social & I wrote a mini-review on this:
academic.oup.com/plcell/artic...
12/🧵
academic.oup.com/plcell/artic...
12/🧵
We never saw PSII arrays within intact chloroplasts. However, we spotted them in chloroplasts that broke open and exposed their thylakoids to buffer. This is a known in vitro phenomenon called “thylakoid restacking”. Thus, interactions between arrays in two membranes seem to drive restacking!🤐
11/🧵
11/🧵

September 25, 2025 at 6:46 PM
We never saw PSII arrays within intact chloroplasts. However, we spotted them in chloroplasts that broke open and exposed their thylakoids to buffer. This is a known in vitro phenomenon called “thylakoid restacking”. Thus, interactions between arrays in two membranes seem to drive restacking!🤐
11/🧵
11/🧵
There have been many studies on PSII arrays, including #CryoET showing that they can stack thylakoid membranes together. The arrays also form in thylakoids from a variety of species, even diatoms. So it isn’t just a “plant thing”🌿. The catch is—all these studies looked at isolated membranes 🤔
10/🧵
10/🧵

September 25, 2025 at 6:39 PM
There have been many studies on PSII arrays, including #CryoET showing that they can stack thylakoid membranes together. The arrays also form in thylakoids from a variety of species, even diatoms. So it isn’t just a “plant thing”🌿. The catch is—all these studies looked at isolated membranes 🤔
10/🧵
10/🧵
But there’s a curious loose end to this story. Scientists have been looking at PSII organization in thylakoids for over 50 years (including some really stunning freeze fracture 🔬😍). Sometimes PSII looks randomly oriented, and other times it forms extended 2D crystalline arrays in the membrane 💠
9/🧵
9/🧵

September 25, 2025 at 6:34 PM
But there’s a curious loose end to this story. Scientists have been looking at PSII organization in thylakoids for over 50 years (including some really stunning freeze fracture 🔬😍). Sometimes PSII looks randomly oriented, and other times it forms extended 2D crystalline arrays in the membrane 💠
9/🧵
9/🧵
We also observed that PSII concentration was uniform throughout each granum, irrespective of distance from the margin. Thus, we propose a simple two-domain model of thylakoid organization in green algae and plants. And yes, we speculate about domain #PhaseSeparation. I await your hate mail 📬🤗
8/🧵
8/🧵

September 25, 2025 at 6:31 PM
We also observed that PSII concentration was uniform throughout each granum, irrespective of distance from the margin. Thus, we propose a simple two-domain model of thylakoid organization in green algae and plants. And yes, we speculate about domain #PhaseSeparation. I await your hate mail 📬🤗
8/🧵
8/🧵
For decades, the idea has persisted that grana margins have a distinct protein composition and provide a zone where PSI & PSII can intermix. However, our "membranograms" revealed strict lateral heterogeneity between PSI & PSII at the margins, just like we previously observed in green algae 🟦🟨
7/🧵
7/🧵

September 25, 2025 at 6:26 PM
For decades, the idea has persisted that grana margins have a distinct protein composition and provide a zone where PSI & PSII can intermix. However, our "membranograms" revealed strict lateral heterogeneity between PSI & PSII at the margins, just like we previously observed in green algae 🟦🟨
7/🧵
7/🧵
Contrary to previous models, our native views showed that grana are not perfect cylinders, but have wavy undulating edges 🌊🏄♀️. Here we slice through a few grana🥞 from the top– Do you see all the little protein densities in the membranes?🔍 Those are photosynthetic complexes. How to analyze them?
5/🧵
5/🧵
September 25, 2025 at 6:14 PM
Contrary to previous models, our native views showed that grana are not perfect cylinders, but have wavy undulating edges 🌊🏄♀️. Here we slice through a few grana🥞 from the top– Do you see all the little protein densities in the membranes?🔍 Those are photosynthetic complexes. How to analyze them?
5/🧵
5/🧵
Our data enabled us to quantify native thylakoid architecture in the grana. Getting precise numbers is important for modeling photosynthetic regulation. Here’s a comparison to similar measurements made by classical TEM. We’d like to apply #CryoET to the dark-to-light💡 transition in the future.
4/🧵
4/🧵

September 25, 2025 at 6:09 PM
Our data enabled us to quantify native thylakoid architecture in the grana. Getting precise numbers is important for modeling photosynthetic regulation. Here’s a comparison to similar measurements made by classical TEM. We’d like to apply #CryoET to the dark-to-light💡 transition in the future.
4/🧵
4/🧵
We made an interesting observation about plastoglobules, lipid-protein storage droplets that share one leaflet with thylakoid membranes. We saw that plastoglobules often sat within little holes in the thylakoids 🟠🕳️, perhaps increasing the area of the contact site, facilitating exchange 🔄
3/🧵
3/🧵

September 25, 2025 at 6:06 PM
We made an interesting observation about plastoglobules, lipid-protein storage droplets that share one leaflet with thylakoid membranes. We saw that plastoglobules often sat within little holes in the thylakoids 🟠🕳️, perhaps increasing the area of the contact site, facilitating exchange 🔄
3/🧵
3/🧵
Unlike in algae and other phototrophs, plant thylakoids form specialized grana stacks🥞 interconnected by single thylakoids called stromal lamellae. The 3D membrane architecture we observed is consistent with the “helical staircase model” proposed from classical TEM (💚🔬 Andrew Staehelin, RIP).
2/🧵
2/🧵

September 25, 2025 at 6:02 PM
Unlike in algae and other phototrophs, plant thylakoids form specialized grana stacks🥞 interconnected by single thylakoids called stromal lamellae. The 3D membrane architecture we observed is consistent with the “helical staircase model” proposed from classical TEM (💚🔬 Andrew Staehelin, RIP).
2/🧵
2/🧵
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
September 25, 2025 at 6:00 PM
Time for a thread!🧵 How different is the molecular organization of thylakoids in “higher” plants🌱? To find out, we teamed up with @profmattjohnson.bsky.social to dive into spinach chloroplasts with #CryoET ❄️🔬. Curious? ..Read on!
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
#TeamTomo #PlantScience 🧪 🧶🧬 🌾
elifesciences.org/articles/105...
1/🧵
Thanks William! Regarding the physiological significance of PSII lattices, please see the "How Fluid are Thylakoid membranes" opinion piece that @wojwie.bsky.social and I wrote here:
academic.oup.com/plcell/artic...
academic.oup.com/plcell/artic...

September 25, 2025 at 10:05 AM
Thanks William! Regarding the physiological significance of PSII lattices, please see the "How Fluid are Thylakoid membranes" opinion piece that @wojwie.bsky.social and I wrote here:
academic.oup.com/plcell/artic...
academic.oup.com/plcell/artic...
Thanks Alister! That Feynman catchphrase is certainly words to live by for #TeamTomo. I actually used to intro my talks with it. But the trick is in the full quote-- these "easy" observations rely on making our microscopes (and tools for analysis) more powerful. Thanks for your help with #MemBrain 🤗

September 20, 2025 at 8:18 PM
Thanks Alister! That Feynman catchphrase is certainly words to live by for #TeamTomo. I actually used to intro my talks with it. But the trick is in the full quote-- these "easy" observations rely on making our microscopes (and tools for analysis) more powerful. Thanks for your help with #MemBrain 🤗
Yes, very carefully worded. The null hypothesis must come first, and it can't be ruled out (but there are significant challenges to it?). It was these paragraphs about the alternative hypothesis that got my heart aflutter😍. And again, I'm NO expert, but this seems fairly well considered... no? 🍎🍏


September 11, 2025 at 2:22 PM
Yes, very carefully worded. The null hypothesis must come first, and it can't be ruled out (but there are significant challenges to it?). It was these paragraphs about the alternative hypothesis that got my heart aflutter😍. And again, I'm NO expert, but this seems fairly well considered... no? 🍎🍏
Thanks @dodonova-sveta.bsky.social for visiting us at the @biozentrum.unibas.ch and giving a fascinating talk on the structures and mysteries of archaeal chromatin. The room was packed!🤓
#ArchaeaSky #TeamTomo 🧪🧶🧬
Check out their lab’s new study on asgard hypernucleosomes: bsky.app/profile/dodo...
#ArchaeaSky #TeamTomo 🧪🧶🧬
Check out their lab’s new study on asgard hypernucleosomes: bsky.app/profile/dodo...

September 9, 2025 at 4:53 PM
Thanks @dodonova-sveta.bsky.social for visiting us at the @biozentrum.unibas.ch and giving a fascinating talk on the structures and mysteries of archaeal chromatin. The room was packed!🤓
#ArchaeaSky #TeamTomo 🧪🧶🧬
Check out their lab’s new study on asgard hypernucleosomes: bsky.app/profile/dodo...
#ArchaeaSky #TeamTomo 🧪🧶🧬
Check out their lab’s new study on asgard hypernucleosomes: bsky.app/profile/dodo...
Theme 1 of the embo.org ESCRT meeting: Tentacles across scales 🐙 …from simulated asgard archaea to swirly filaments 🌀
Andela Saric, Adai Colom, Qing-Tao Shen, John McCullough 🧶🧬🧪
Andela Saric, Adai Colom, Qing-Tao Shen, John McCullough 🧶🧬🧪



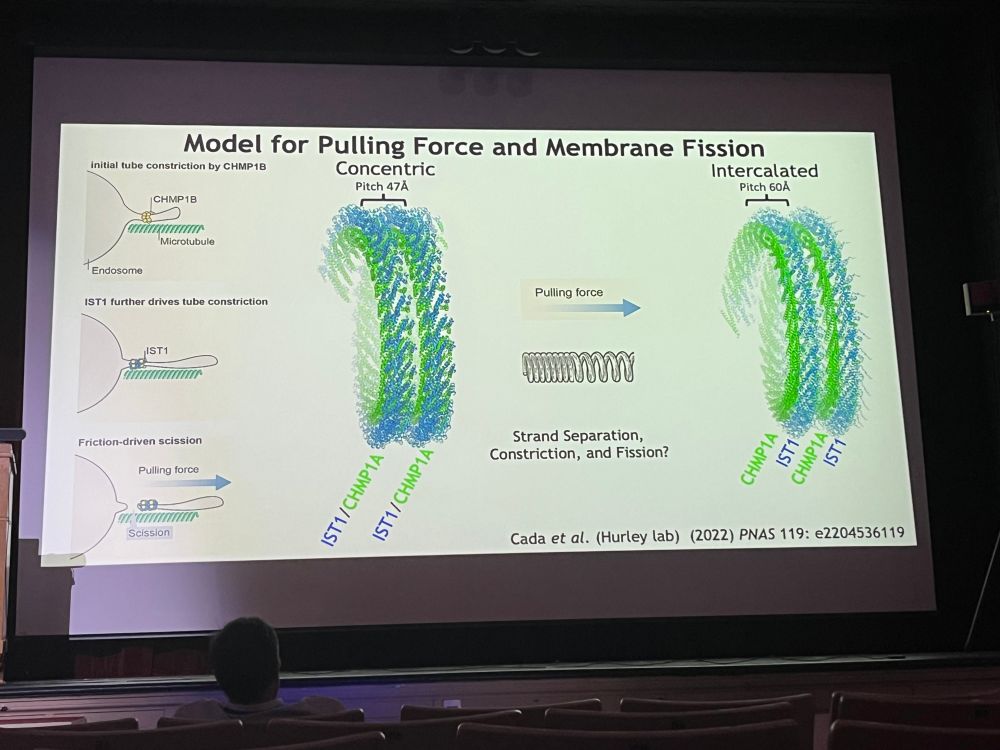
June 27, 2025 at 6:40 PM
Theme 1 of the embo.org ESCRT meeting: Tentacles across scales 🐙 …from simulated asgard archaea to swirly filaments 🌀
Andela Saric, Adai Colom, Qing-Tao Shen, John McCullough 🧶🧬🧪
Andela Saric, Adai Colom, Qing-Tao Shen, John McCullough 🧶🧬🧪
The @embo.org ESCRT meeting was packed with membrane remodeling from every branch of the tree of life🌳. Some awesome science 🧪 highlights in the next posts 🧵. But a first impression… the venue!! Wow, Caux castle up a cogwheel train, overlooking Lac Léman! Perfect! 🌄 Thank you @rouxlab.bsky.social 🤗




June 27, 2025 at 2:54 PM
The @embo.org ESCRT meeting was packed with membrane remodeling from every branch of the tree of life🌳. Some awesome science 🧪 highlights in the next posts 🧵. But a first impression… the venue!! Wow, Caux castle up a cogwheel train, overlooking Lac Léman! Perfect! 🌄 Thank you @rouxlab.bsky.social 🤗
Such a joy to host @gaiapigino.bsky.social as the keynote speaker of our annual @biozentrum.unibas.ch @unibas.ch symposium! It’s amazing how far Gaia and her team have pushed the IFT field. Nice exchange with @computingcaitie.bsky.social and the rest of our group. Thanks for visiting! #TeamTomo 🧪🧶🧬🔬




June 12, 2025 at 9:59 PM
Such a joy to host @gaiapigino.bsky.social as the keynote speaker of our annual @biozentrum.unibas.ch @unibas.ch symposium! It’s amazing how far Gaia and her team have pushed the IFT field. Nice exchange with @computingcaitie.bsky.social and the rest of our group. Thanks for visiting! #TeamTomo 🧪🧶🧬🔬
And from the other direction (not mirrored I hope 🤚✋). Thanks so much for hosting us. So, Basel last week and Paris this week…. where’s dinner next week? (For those with keen eyes: there’s another member of #TeamTomo in this photo. But who?🔎)

May 19, 2025 at 9:38 PM
And from the other direction (not mirrored I hope 🤚✋). Thanks so much for hosting us. So, Basel last week and Paris this week…. where’s dinner next week? (For those with keen eyes: there’s another member of #TeamTomo in this photo. But who?🔎)

