Brian Kelch
@briankelch.bsky.social
My lab at UMass Chan Medical School studies virus assembly and DNA replication/repair using structural biology, biophysics, and biochemistry. Habitual Line-Stepper.
umassmed.edu/kelchlab
umassmed.edu/kelchlab
RFC is conceptually analogous to a GTPase, which has dedicated Nucleotide Exchange Factors and NTPase Activating Partners. What makes RFC unique is that its own substrates are the NEF (PCNA) and NAP (DNA). (13/16)

July 7, 2025 at 7:06 PM
RFC is conceptually analogous to a GTPase, which has dedicated Nucleotide Exchange Factors and NTPase Activating Partners. What makes RFC unique is that its own substrates are the NEF (PCNA) and NAP (DNA). (13/16)
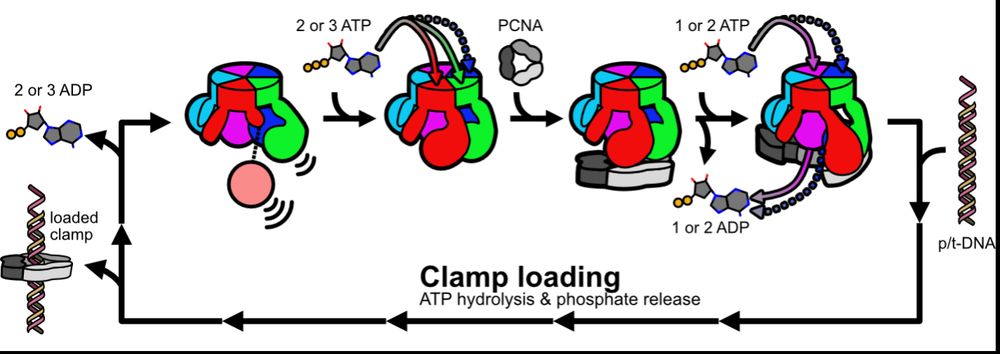
Based on our data, we propose a model whereby fast ATP binding to some subunits readies RFC to bind PCNA, and then PCNA completes RFC’s nucleotide exchange en route to clamp loading. We propose this prevents futile ATP hydrolysis and helps ensure RFC binds PCNA before DNA. (12/16)
July 7, 2025 at 7:06 PM
Based on our data, we propose a model whereby fast ATP binding to some subunits readies RFC to bind PCNA, and then PCNA completes RFC’s nucleotide exchange en route to clamp loading. We propose this prevents futile ATP hydrolysis and helps ensure RFC binds PCNA before DNA. (12/16)
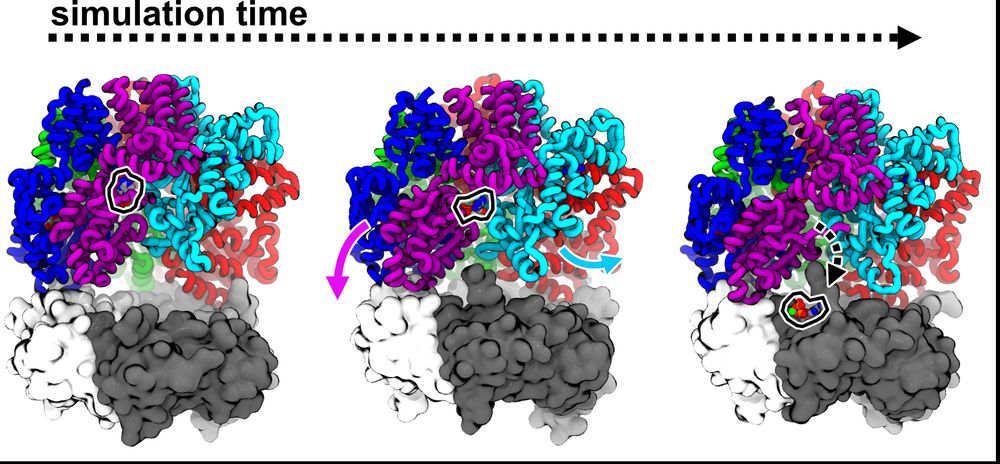
Josh performed more MD sims to try to understand how PCNA could promote nucleotide exchange in RFC. His sims predict that the D/E interface weakly bridged by ADP can be pried open as D binds PCNA, accelerating release. (11/16)
July 7, 2025 at 7:06 PM
Josh performed more MD sims to try to understand how PCNA could promote nucleotide exchange in RFC. His sims predict that the D/E interface weakly bridged by ADP can be pried open as D binds PCNA, accelerating release. (11/16)
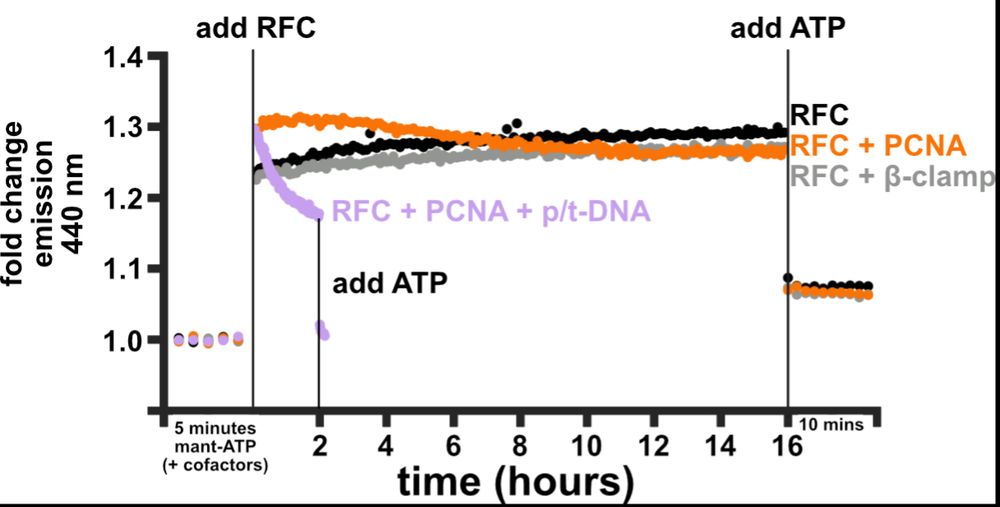
So, what catalyzes nucleotide exchange? Josh developed a FRET assay using a labeled ATP analog to monitor ATP binding to RFC. RFC alone binds some ATPs fast but then binds more ATP slowly, consistent with A&B (and maybe C) exchanging faster than D. PCNA speeds up these slow binding steps! (9/16)
July 7, 2025 at 7:06 PM
So, what catalyzes nucleotide exchange? Josh developed a FRET assay using a labeled ATP analog to monitor ATP binding to RFC. RFC alone binds some ATPs fast but then binds more ATP slowly, consistent with A&B (and maybe C) exchanging faster than D. PCNA speeds up these slow binding steps! (9/16)
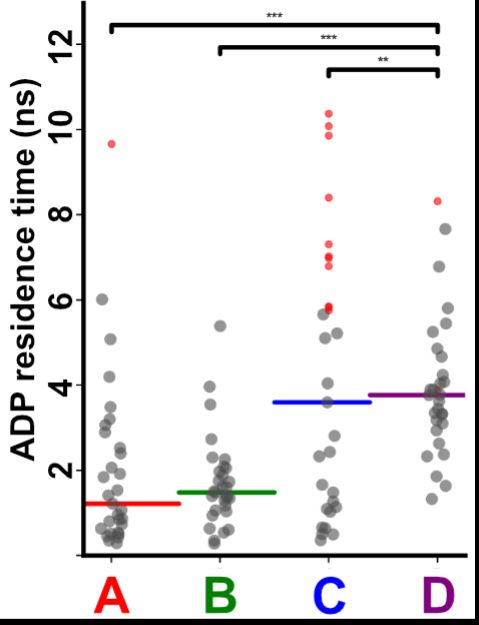
To test how interface opening plays a role in ADP release, Josh performed tRAMD(doi.org/10.1021/acs....), which is a technique that predicts relative release rates. He found that the half-off rate for C or D is slower than A or B, consistent with his cryo-EM. (8/16)
July 7, 2025 at 7:06 PM
To test how interface opening plays a role in ADP release, Josh performed tRAMD(doi.org/10.1021/acs....), which is a technique that predicts relative release rates. He found that the half-off rate for C or D is slower than A or B, consistent with his cryo-EM. (8/16)
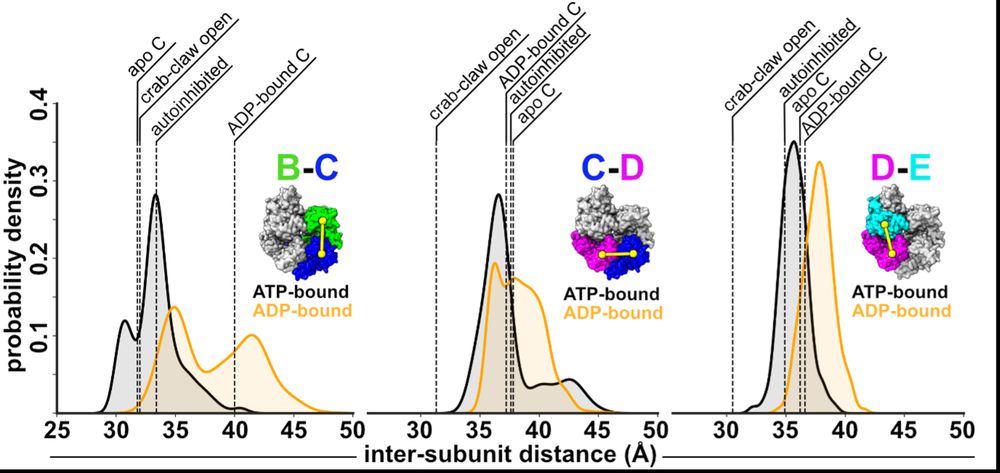
Next, we wondered how the dynamics of RFC influence ADP release? So, Josh performed MD sims. His sims predict that ATP-interfaces are tighter than ADP ones, and that the B/C interface can open farthest, offering an explanation why this interface is apo in our cryo-EM reconstructions. (7/16)
July 7, 2025 at 7:06 PM
Next, we wondered how the dynamics of RFC influence ADP release? So, Josh performed MD sims. His sims predict that ATP-interfaces are tighter than ADP ones, and that the B/C interface can open farthest, offering an explanation why this interface is apo in our cryo-EM reconstructions. (7/16)
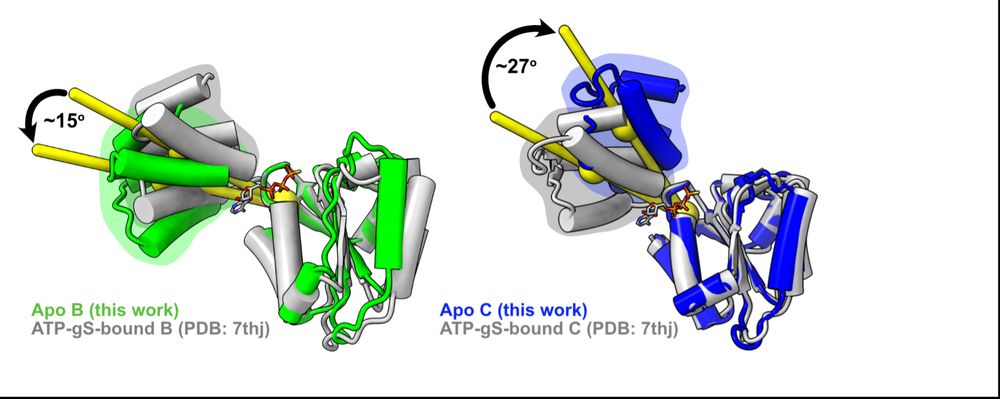
First, we looked at how RFC subunits respond to nucleotide binding by comparing Josh’s apo subunits to our lab’s previous ATPgS-bound subunits and found that the B&C subunits’ lids rotate in opposite directions! We had never seen this in a AAA+ before (if you know of any, please let us know!) (6/16)
July 7, 2025 at 7:06 PM
First, we looked at how RFC subunits respond to nucleotide binding by comparing Josh’s apo subunits to our lab’s previous ATPgS-bound subunits and found that the B&C subunits’ lids rotate in opposite directions! We had never seen this in a AAA+ before (if you know of any, please let us know!) (6/16)
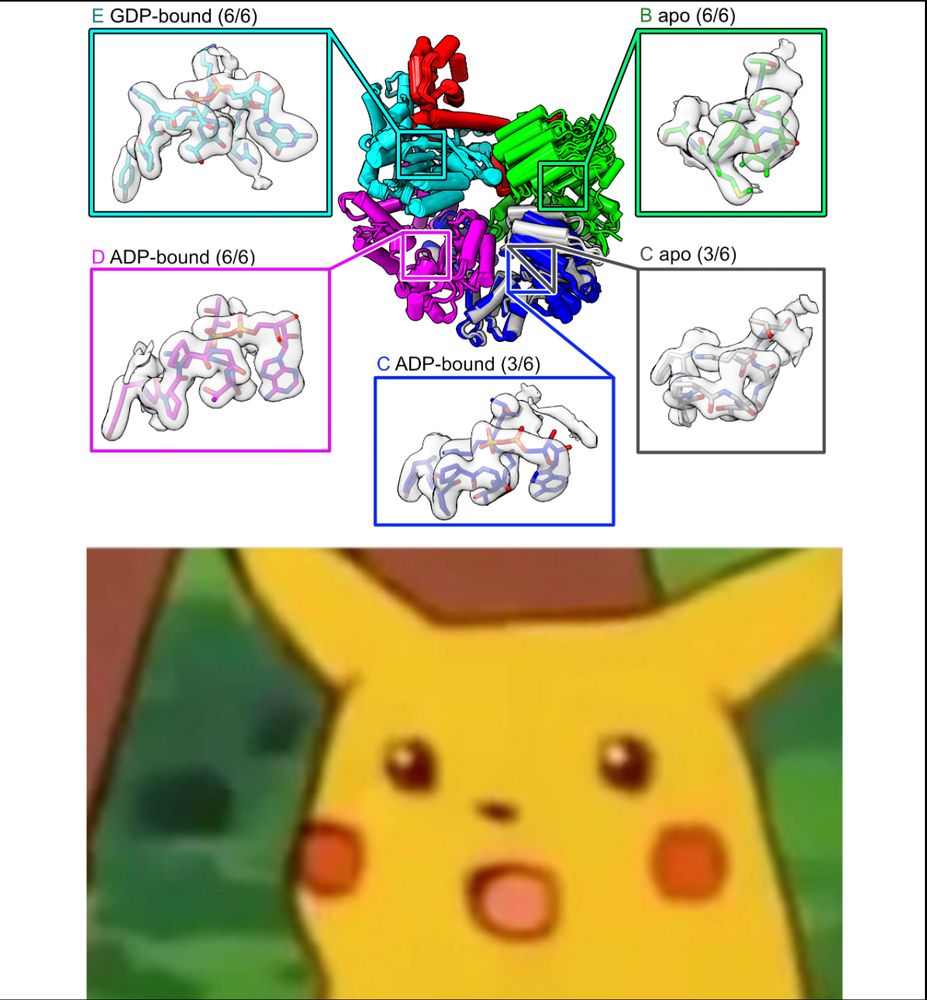
Imagine our surprise when we saw that the C and D subunits were ADP-bound! Because he never added nucleotide, these ADPs must have co-purified with RFC over a 3 day prep, meaning that RFC releases ADP very slowly from these subunits. (4/16)
July 7, 2025 at 7:06 PM
Imagine our surprise when we saw that the C and D subunits were ADP-bound! Because he never added nucleotide, these ADPs must have co-purified with RFC over a 3 day prep, meaning that RFC releases ADP very slowly from these subunits. (4/16)
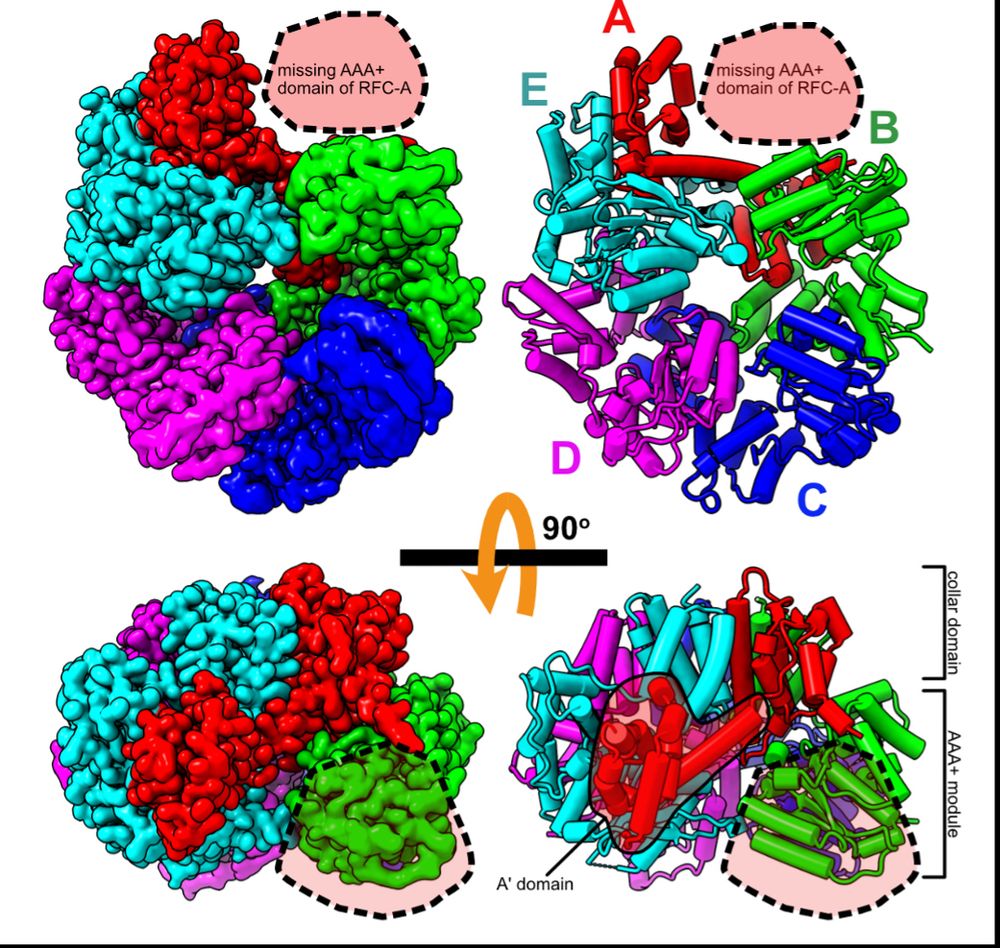
To get insights into RFC recycling, postdoc @joshuapajak.bsky.social determined the structure of RFC in the apo state. He got great overall resolution of the core RFC complex, but the AAA+ domain of the A subunit is missing, suggesting it is flexibly tethered when apo. (3/16)
July 7, 2025 at 7:06 PM
To get insights into RFC recycling, postdoc @joshuapajak.bsky.social determined the structure of RFC in the apo state. He got great overall resolution of the core RFC complex, but the AAA+ domain of the A subunit is missing, suggesting it is flexibly tethered when apo. (3/16)
Recently, our lab showed how an ATP-bound RFC loads PCNA onto DNA during replication and repair (doi.org/10.7554/eLif...). But RFC loads PCNA extremely fast, so that when RFC has done its job once, how does it prep for the next round? (2/16)

July 7, 2025 at 7:06 PM
Recently, our lab showed how an ATP-bound RFC loads PCNA onto DNA during replication and repair (doi.org/10.7554/eLif...). But RFC loads PCNA extremely fast, so that when RFC has done its job once, how does it prep for the next round? (2/16)
Every day we make enough DNA to go to the moon and back. So, all our DNA replication enzymes work fast, right? In our latest preprint, we show that a key enzyme, the clamp loader ATPase RFC, releases ADP much slower than it loads PCNA. What gives? (1/16)
doi.org/10.1101/2025...
doi.org/10.1101/2025...

July 7, 2025 at 7:06 PM
Every day we make enough DNA to go to the moon and back. So, all our DNA replication enzymes work fast, right? In our latest preprint, we show that a key enzyme, the clamp loader ATPase RFC, releases ADP much slower than it loads PCNA. What gives? (1/16)
doi.org/10.1101/2025...
doi.org/10.1101/2025...
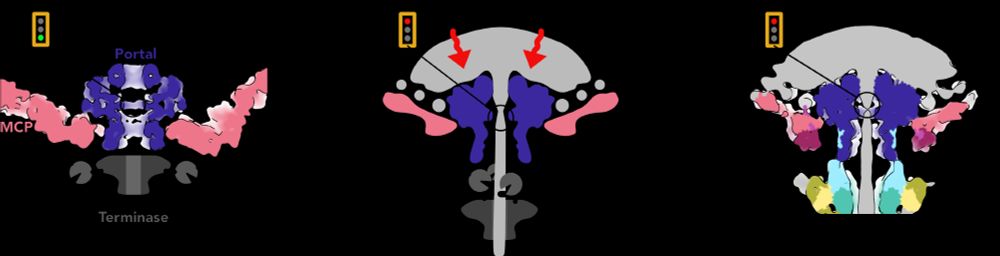
Putting that together with decades of research into phage genome packaging, we propose a speculative model for how the Portal switches from DNA-pumping mode to Tail attachment mode, using the pressure-induced piston motion of Portal to displace the motor. /12
April 18, 2025 at 2:22 PM
Putting that together with decades of research into phage genome packaging, we propose a speculative model for how the Portal switches from DNA-pumping mode to Tail attachment mode, using the pressure-induced piston motion of Portal to displace the motor. /12
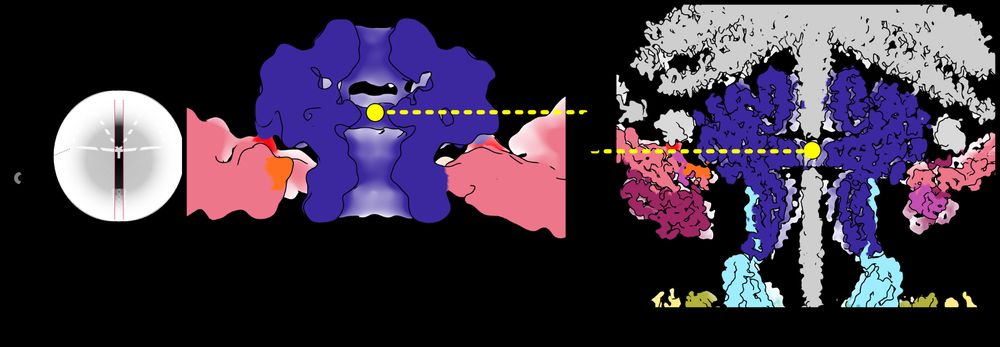
So how does the Portal know it’s time to close the pore? We got a clue by comparing the position of the Portal in capsids before and after DNA has been pumped in. The pressure from DNA pushes the Portal lower in the capsid aperture like a piston! /11
April 18, 2025 at 2:20 PM
So how does the Portal know it’s time to close the pore? We got a clue by comparing the position of the Portal in capsids before and after DNA has been pumped in. The pressure from DNA pushes the Portal lower in the capsid aperture like a piston! /11
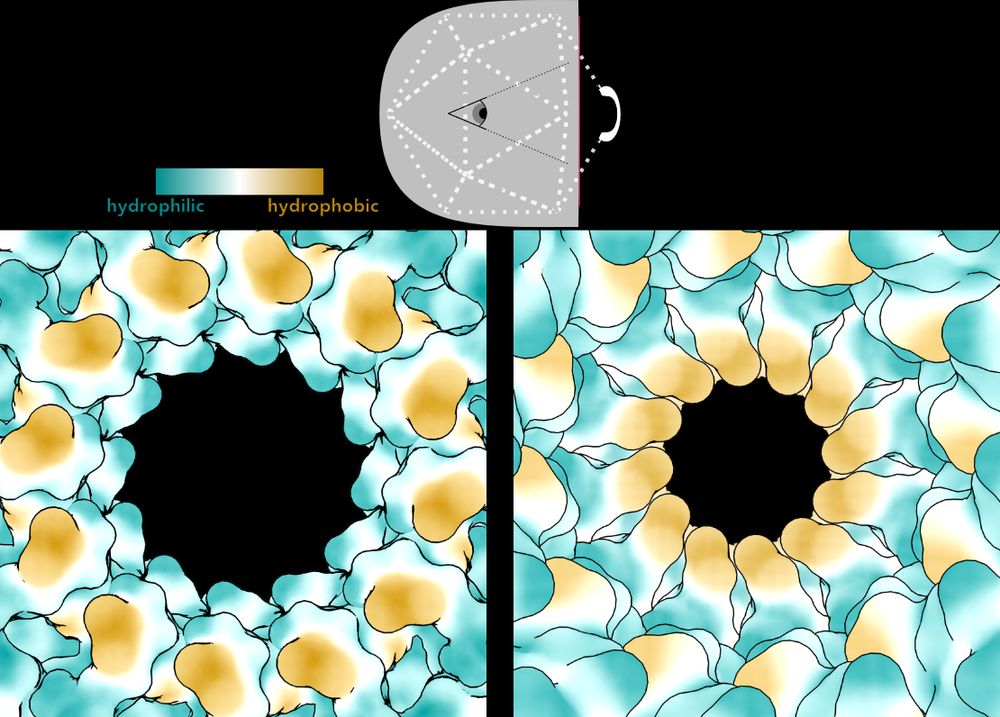
On the left, Bayfield et al. showed the pore through Portal in an early, immature capsid. A specialized motor pumps the genome into the capsid through this open, hydrophilic pore. Then, it constricts in the mature phage, becoming narrow and hydrophobic. /10
April 18, 2025 at 2:19 PM
On the left, Bayfield et al. showed the pore through Portal in an early, immature capsid. A specialized motor pumps the genome into the capsid through this open, hydrophilic pore. Then, it constricts in the mature phage, becoming narrow and hydrophobic. /10
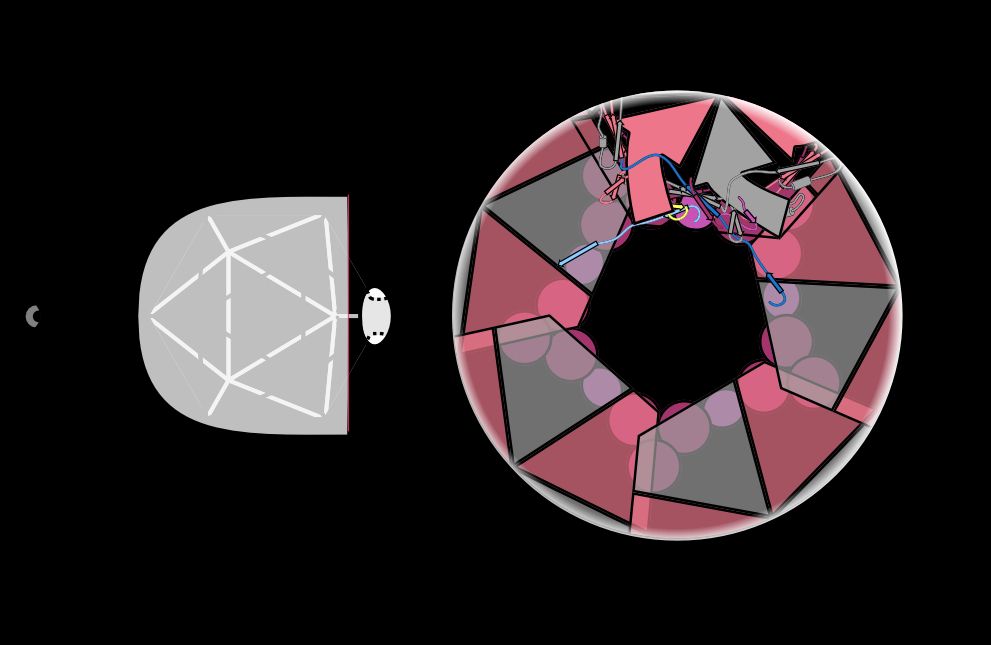
In 2020, Ollie Bayfield and Fred Antson predicted that one component of the Neck complex, the Portal protein (dark blue), would be constricted to hold DNA in the capsid of this phage. And they were right! /9
Bayfield et al elifesciences.org/articles/55517
Bayfield et al elifesciences.org/articles/55517
April 18, 2025 at 2:18 PM
In 2020, Ollie Bayfield and Fred Antson predicted that one component of the Neck complex, the Portal protein (dark blue), would be constricted to hold DNA in the capsid of this phage. And they were right! /9
Bayfield et al elifesciences.org/articles/55517
Bayfield et al elifesciences.org/articles/55517
We know that the Neck of this phage is sufficient to hold its DNA inside the capsid because we also isolated phage that have broken tails, and therefore nothing left but the Neck to retain DNA. /8
April 18, 2025 at 2:17 PM
We know that the Neck of this phage is sufficient to hold its DNA inside the capsid because we also isolated phage that have broken tails, and therefore nothing left but the Neck to retain DNA. /8
But the phage has another problem—it needs a channel through the Neck and Tail to inject its genome into the host when it infects, but it can’t shoot its shot until it finds the right host. How does it keep that channel closed? /7

April 18, 2025 at 2:16 PM
But the phage has another problem—it needs a channel through the Neck and Tail to inject its genome into the host when it infects, but it can’t shoot its shot until it finds the right host. How does it keep that channel closed? /7
Instead of beefing up electrostatic or hydrophobic interactions between these proteins, the phage intertwines different subunits to create topological linkages. This is the same strategy used to make a wicker basket strong! /6

April 18, 2025 at 2:15 PM
Instead of beefing up electrostatic or hydrophobic interactions between these proteins, the phage intertwines different subunits to create topological linkages. This is the same strategy used to make a wicker basket strong! /6
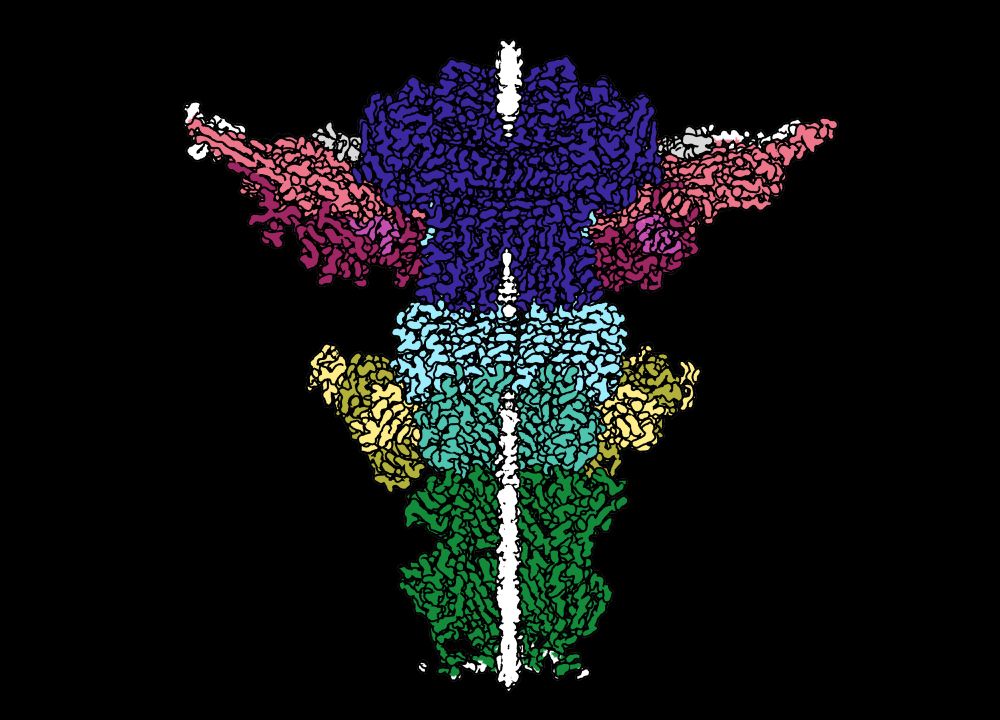
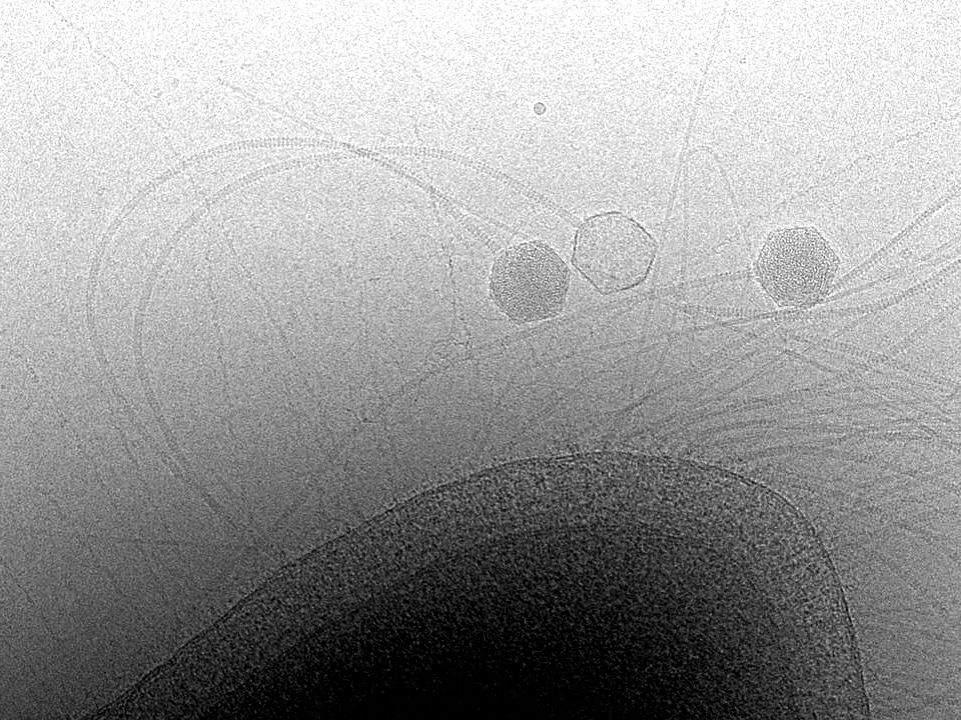
With 105 protein chains and 5 DNA segments, across 4 different symmetries, this is the largest structure the Kelch Lab has ever built (2.4 MDa)! The proteins in pinks/reds are part of the capsid shell, and they form an aperture that the rest of the Neck sits in. /4

April 18, 2025 at 2:13 PM
With 105 protein chains and 5 DNA segments, across 4 different symmetries, this is the largest structure the Kelch Lab has ever built (2.4 MDa)! The proteins in pinks/reds are part of the capsid shell, and they form an aperture that the rest of the Neck sits in. /4
Postdoc @emma.sedivy.bsky.social determined the structure of the Neck complex that joins the capsid head to the tail. We only knew the identity of 4 out of 8 of the proteins in the Neck, but we were able to identify the remaining 4 using ModelAngelo by @kjamali.bsky.social. /3

April 18, 2025 at 2:12 PM
Postdoc @emma.sedivy.bsky.social determined the structure of the Neck complex that joins the capsid head to the tail. We only knew the identity of 4 out of 8 of the proteins in the Neck, but we were able to identify the remaining 4 using ModelAngelo by @kjamali.bsky.social. /3
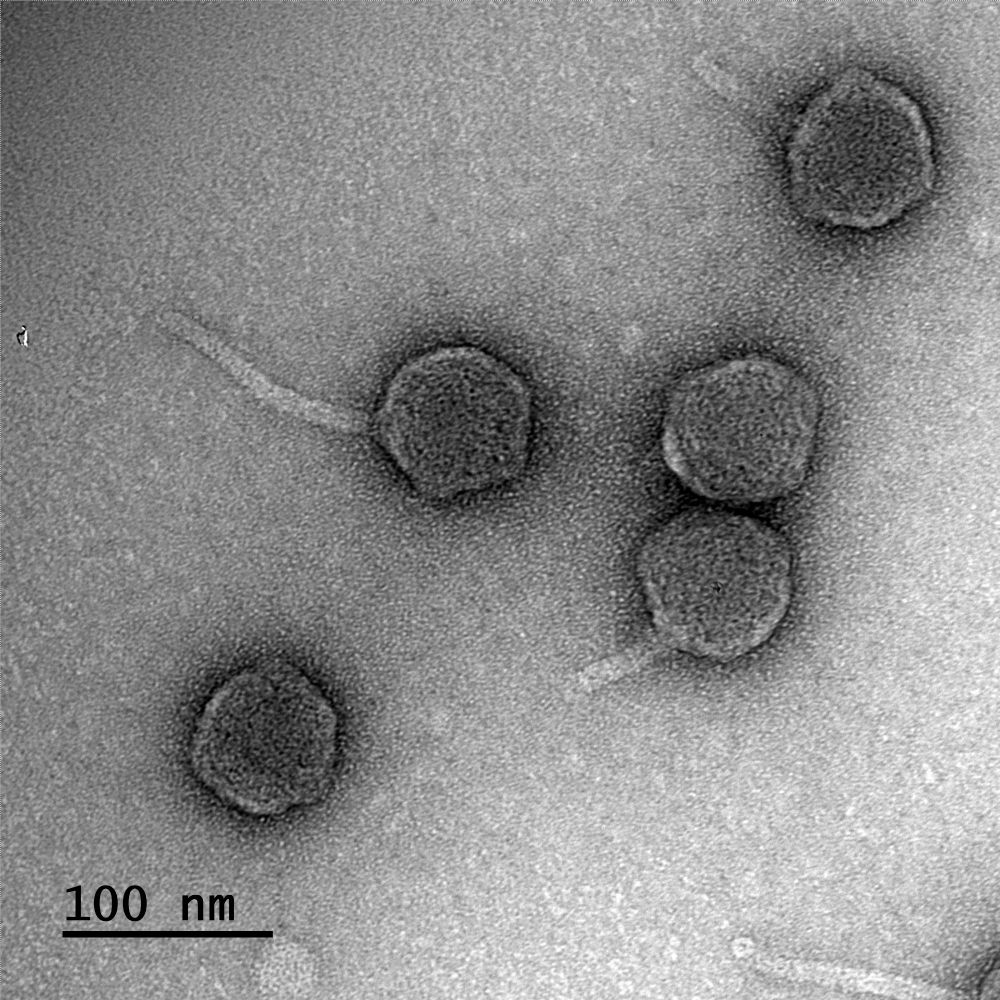
This phage not only lives in hot springs but also has the longest tail of any known phage. Here you see 3 viruses infecting their host, T. thermophilus. One has released its genome into the host, but two still have DNA-filled heads. /2
April 18, 2025 at 2:10 PM
This phage not only lives in hot springs but also has the longest tail of any known phage. Here you see 3 viruses infecting their host, T. thermophilus. One has released its genome into the host, but two still have DNA-filled heads. /2
Did you know that there is so much DNA packed inside a phage capsid that the pressure is 10X higher than in a bottle of champagne? In our latest preprint, we wondered how that is contained in a phage that lives at extremely high temperatures. /1
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
April 18, 2025 at 2:09 PM
Did you know that there is so much DNA packed inside a phage capsid that the pressure is 10X higher than in a bottle of champagne? In our latest preprint, we wondered how that is contained in a phage that lives at extremely high temperatures. /1
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
This leads to a mechanistic hypothesis that Copia and Arc antagonize each others’ capsid assembly by competing for RNA. The functional relevance of Copia in synaptic dev also suggests that we might be watching Copia in process of ‘domestication’. /6

January 4, 2024 at 6:27 PM
This leads to a mechanistic hypothesis that Copia and Arc antagonize each others’ capsid assembly by competing for RNA. The functional relevance of Copia in synaptic dev also suggests that we might be watching Copia in process of ‘domestication’. /6
Amazingly, knockdown of Copia results in increased bouton development and synaptic plasticity, the opposite of an Arc knockout. Copia and Arc are antagonizing each other. 5/

January 4, 2024 at 6:25 PM
Amazingly, knockdown of Copia results in increased bouton development and synaptic plasticity, the opposite of an Arc knockout. Copia and Arc are antagonizing each other. 5/
We find that Copia requires RNA for proper assembly of the capsid structure. The structure shows pores that presumably allow small molecules to enter the capsid, presumably for reverse transcriptase activity. Arc lacks these pores. /4

January 4, 2024 at 6:24 PM
We find that Copia requires RNA for proper assembly of the capsid structure. The structure shows pores that presumably allow small molecules to enter the capsid, presumably for reverse transcriptase activity. Arc lacks these pores. /4

