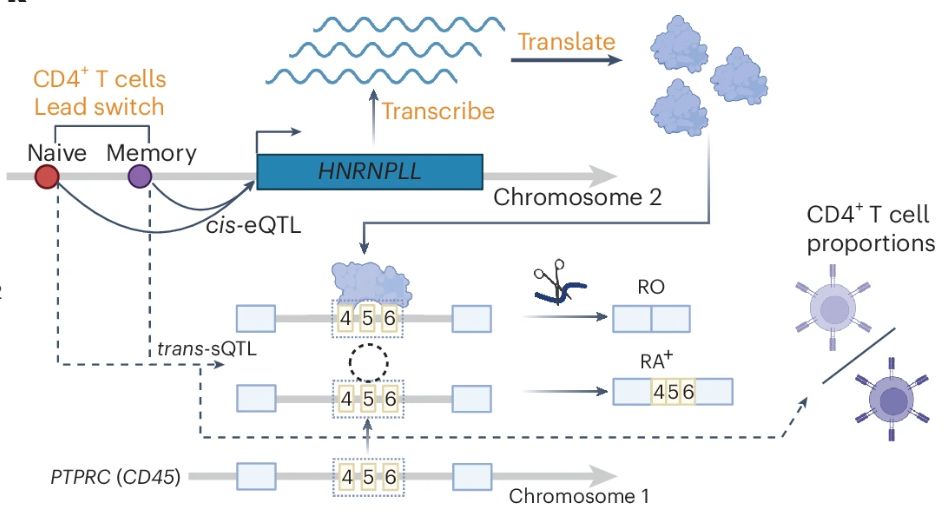
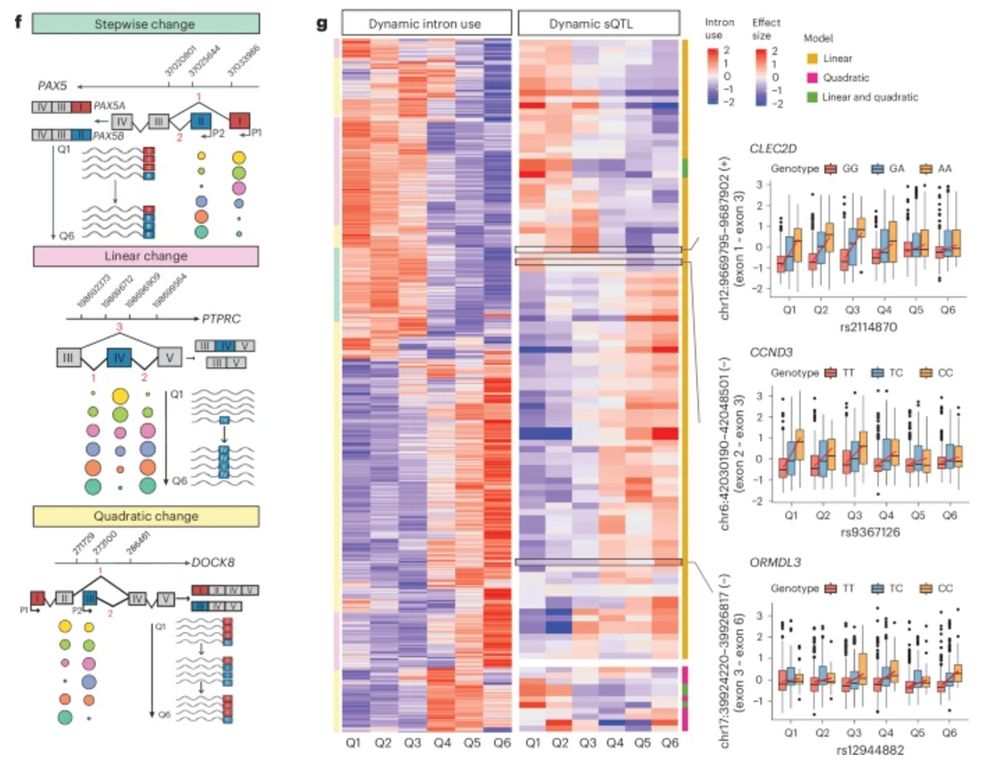
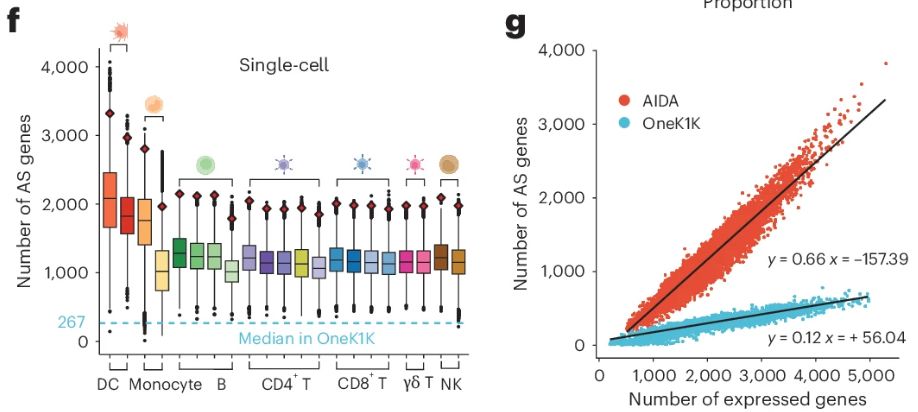
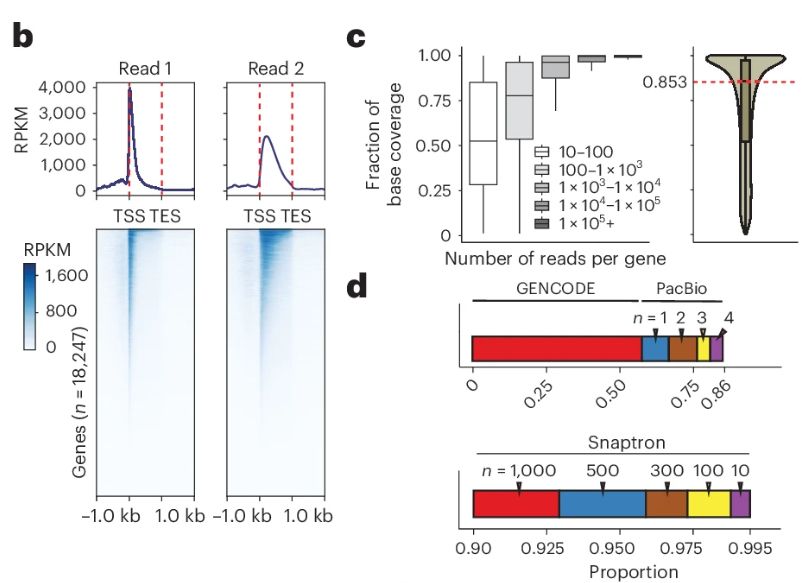
A genetic variant influencing the TCHP exon 4 inclusion, which putatively affects Graves' disease risk. We validated the variant-to-splicing relationship using a minigene assay.
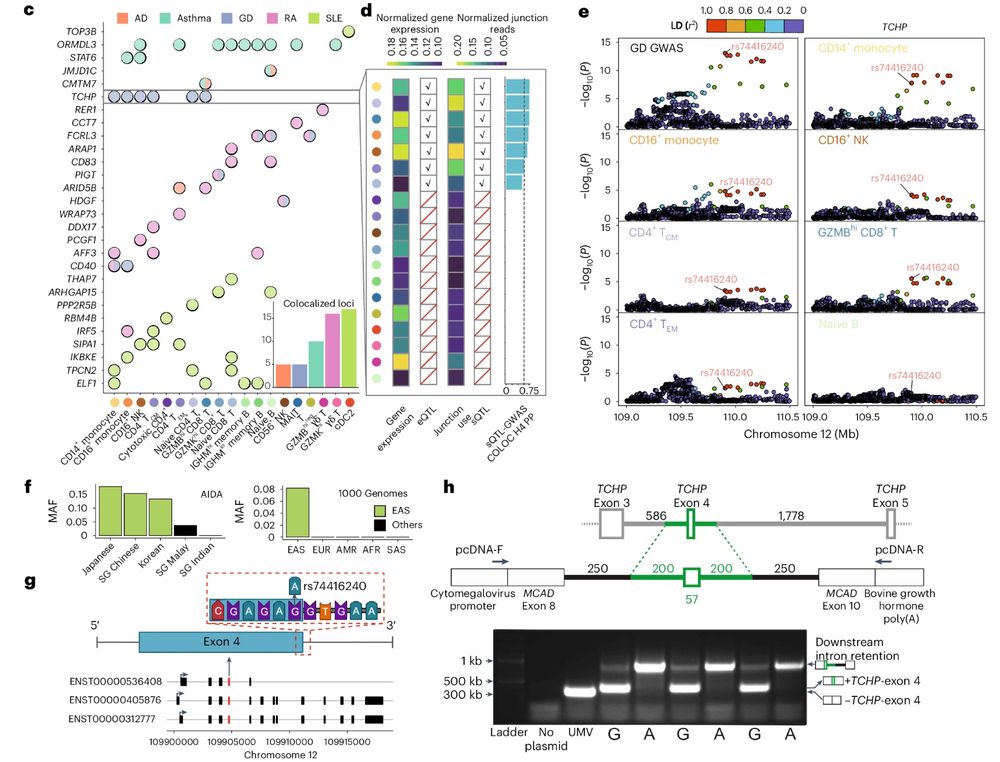
A genetic variant influencing the TCHP exon 4 inclusion, which putatively affects Graves' disease risk. We validated the variant-to-splicing relationship using a minigene assay.