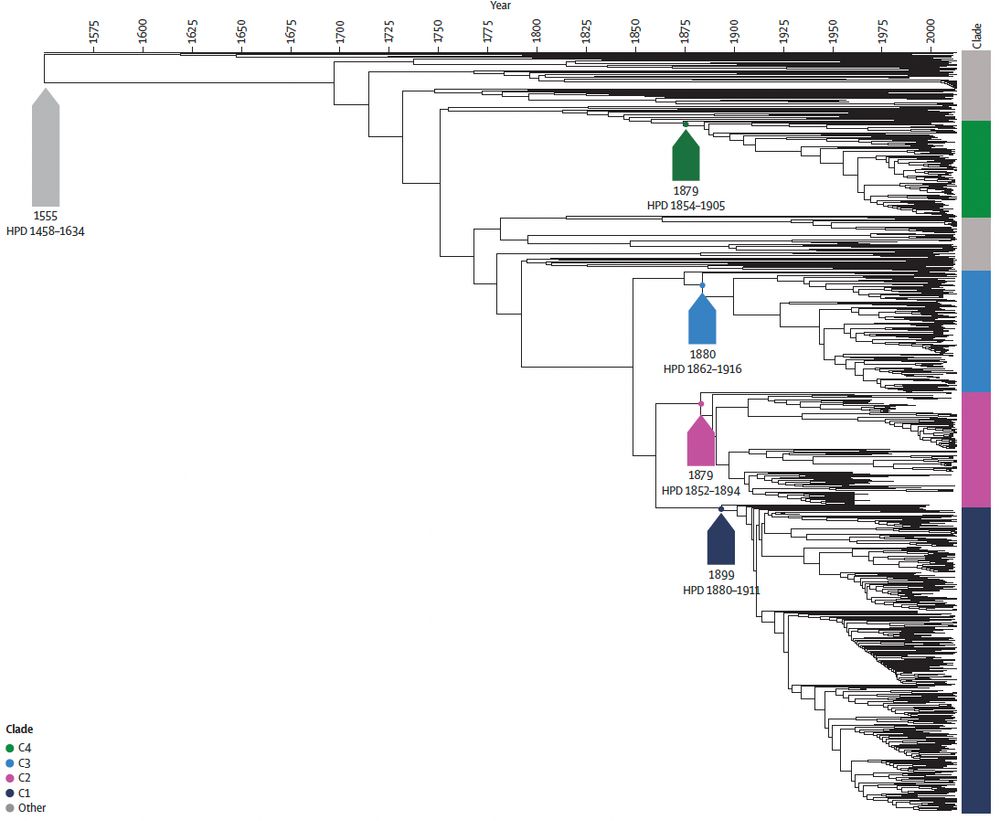
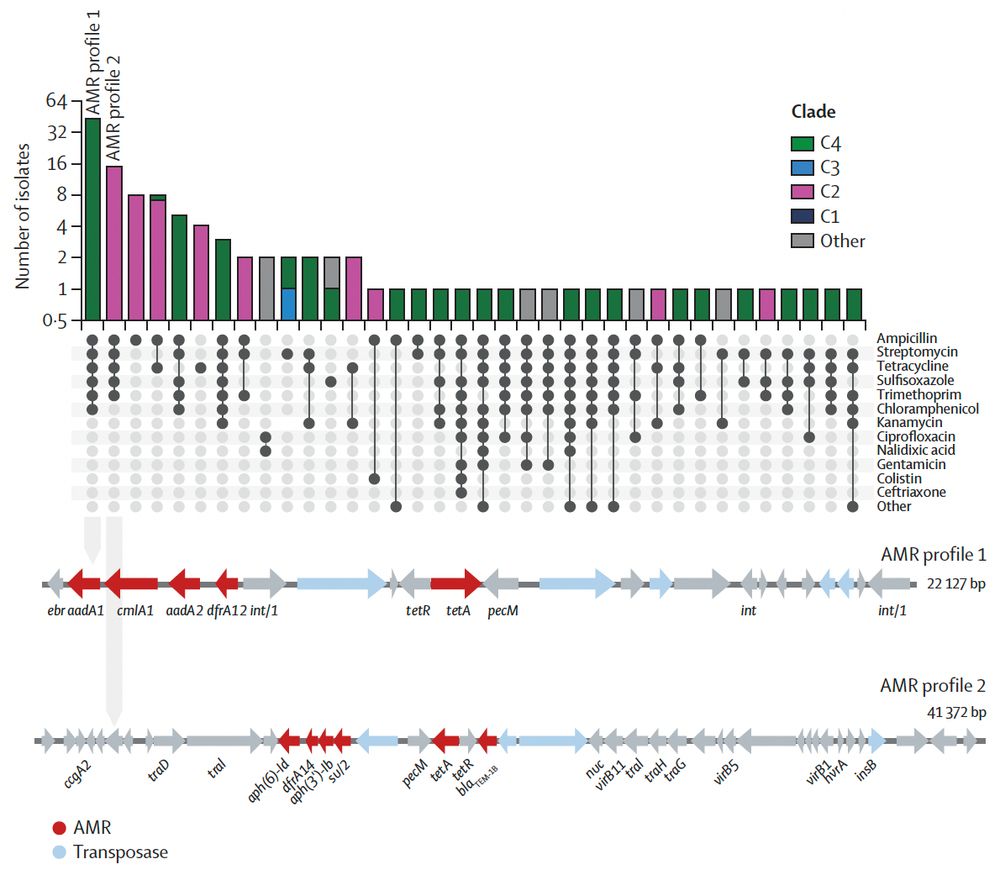
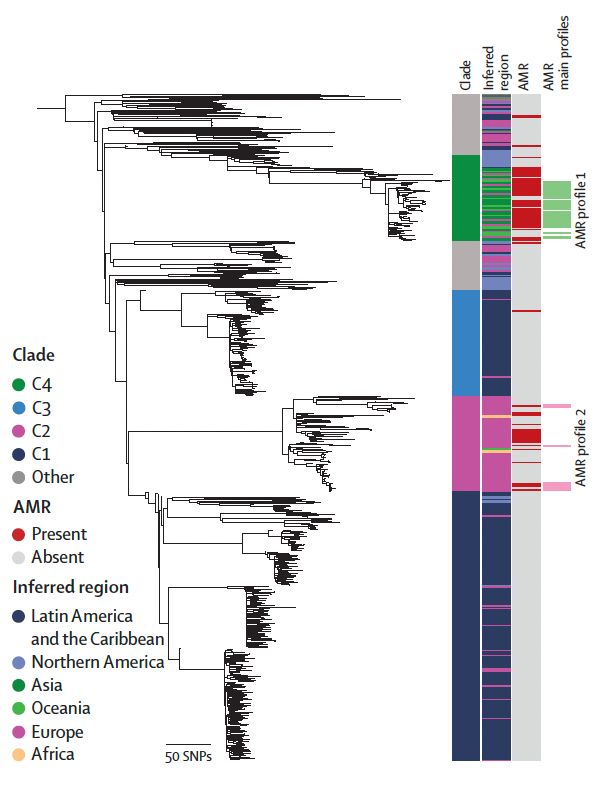
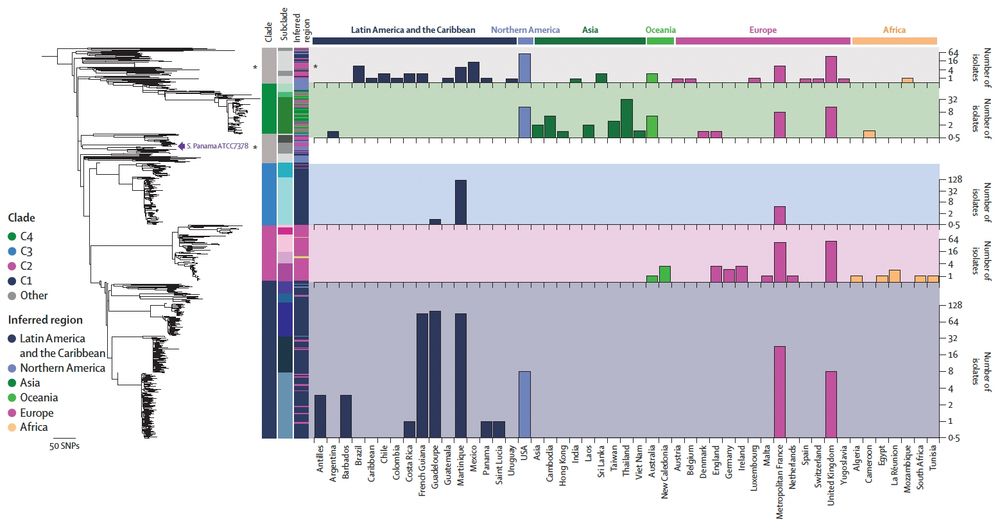
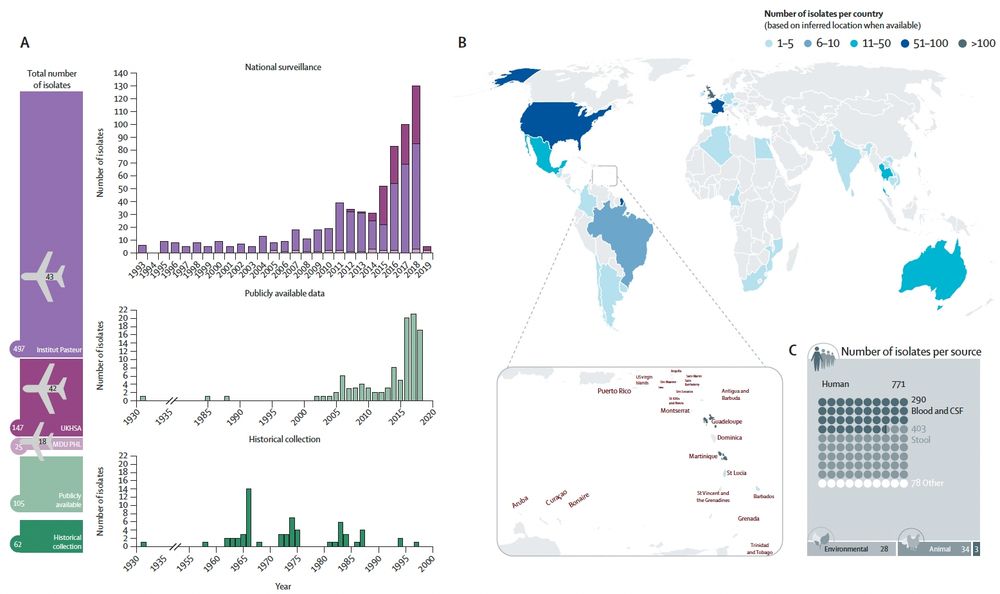

Thank you to @nihr.bsky.social for funding via the second HPRU-GI @livuni-ives.bsky.social.

Thank you to @nihr.bsky.social for funding via the second HPRU-GI @livuni-ives.bsky.social.
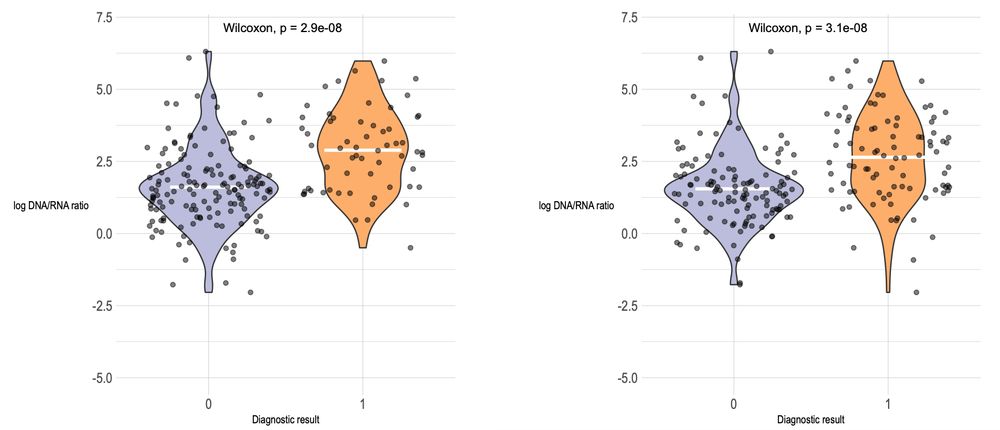
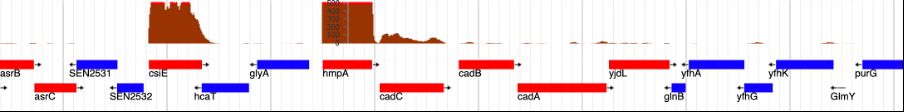
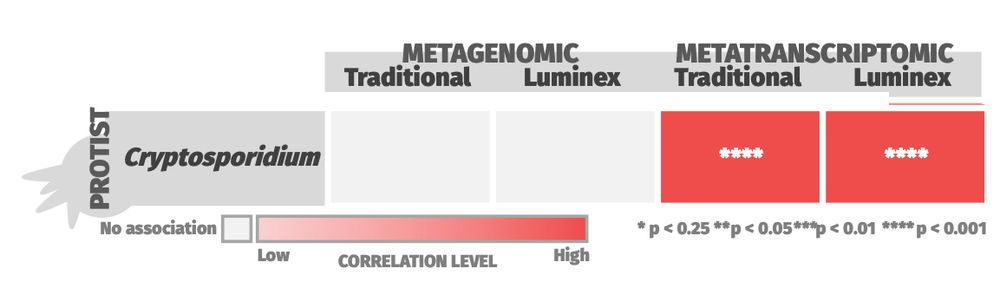
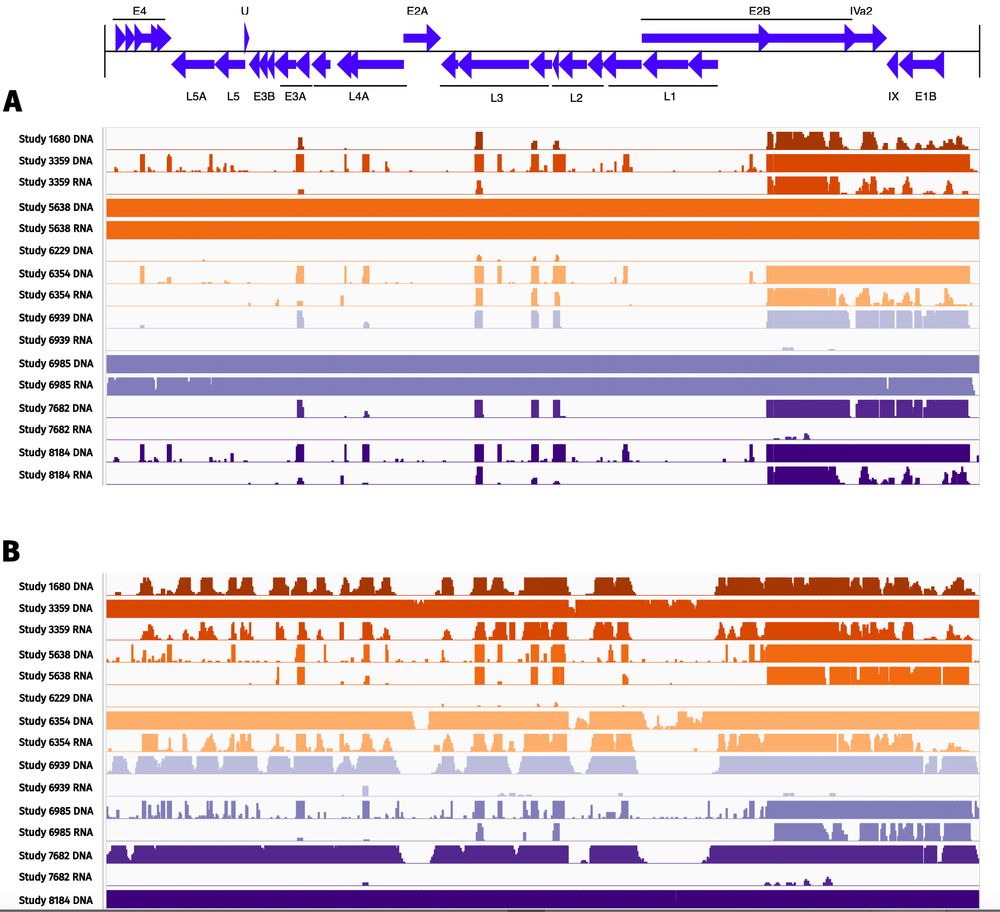
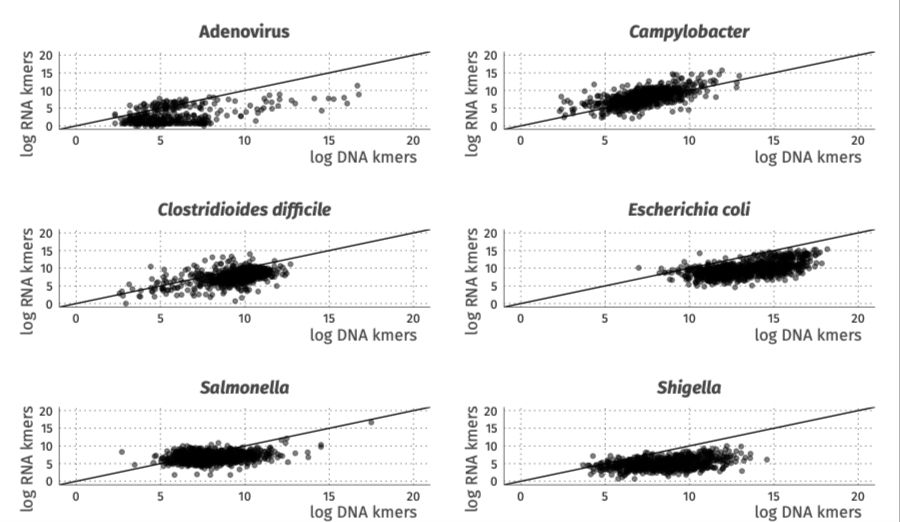
🧪 Metatranscriptomics showed superior sensitivity, with strong correlation to conventional diagnostics for 6/15 pathogens and to Luminex for 8/14.
🧬 Metagenomics performed well for some targets but showed lower overall concordance.
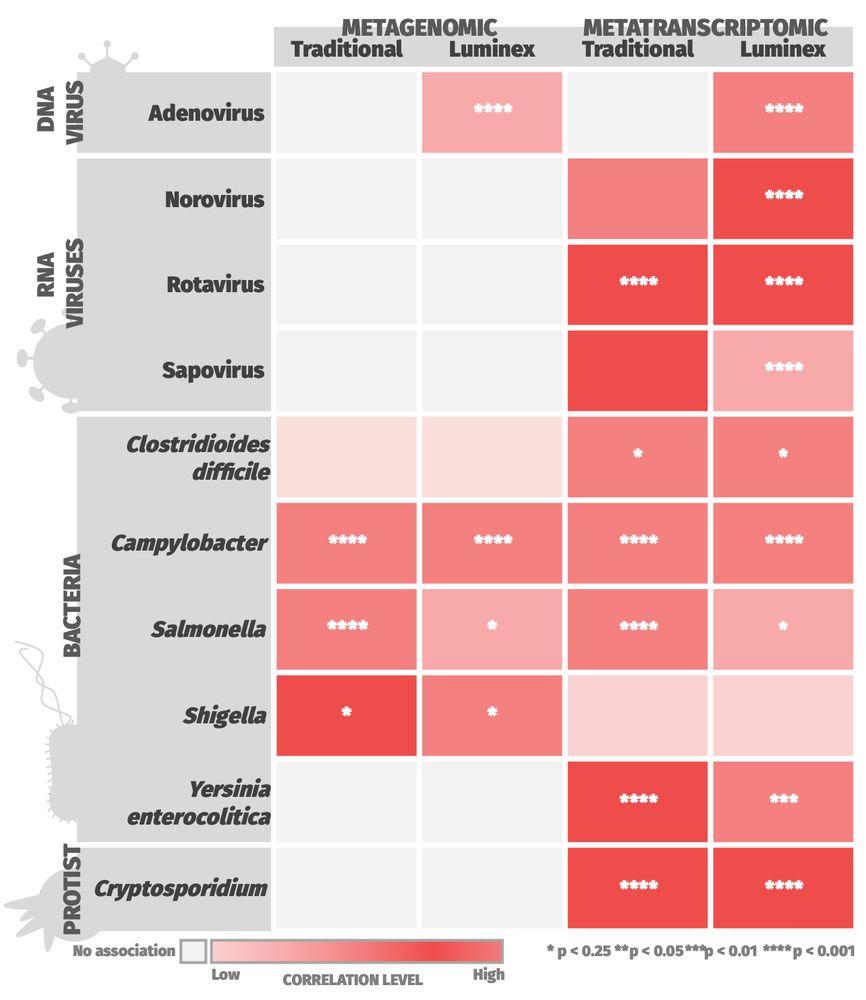
🧪 Metatranscriptomics showed superior sensitivity, with strong correlation to conventional diagnostics for 6/15 pathogens and to Luminex for 8/14.
🧬 Metagenomics performed well for some targets but showed lower overall concordance.