Genome editing / Protein eng. / Molecular medicine 🇨🇦🧬
Kayden-Lambert MGH Research Scholar '23-28
Crescendo: KJ's Dad on watching 🏈 with his son: "This is what we thought we would never get". ❤️👏

Crescendo: KJ's Dad on watching 🏈 with his son: "This is what we thought we would never get". ❤️👏
Really well deserved recognition for an innovator and emerging leader in our field! 👏 🙌
@mgbresearch.bsky.social @harvardmed.bsky.social

Really well deserved recognition for an innovator and emerging leader in our field! 👏 🙌
@mgbresearch.bsky.social @harvardmed.bsky.social








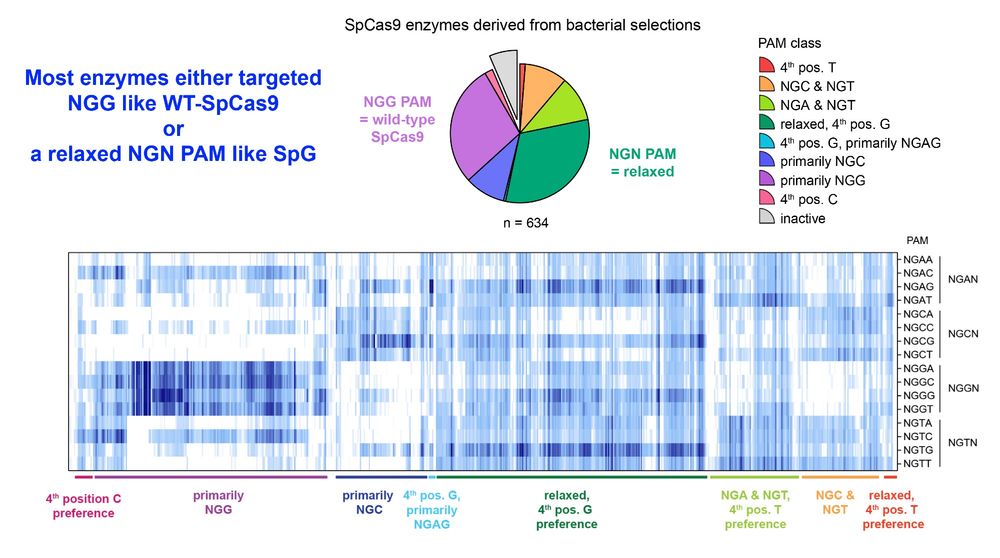
Importantly, PAMmla reversed the biology of Cas9, now able distinguish against a canonical NGGG PAM & instead effectively target an NGTG PAM.
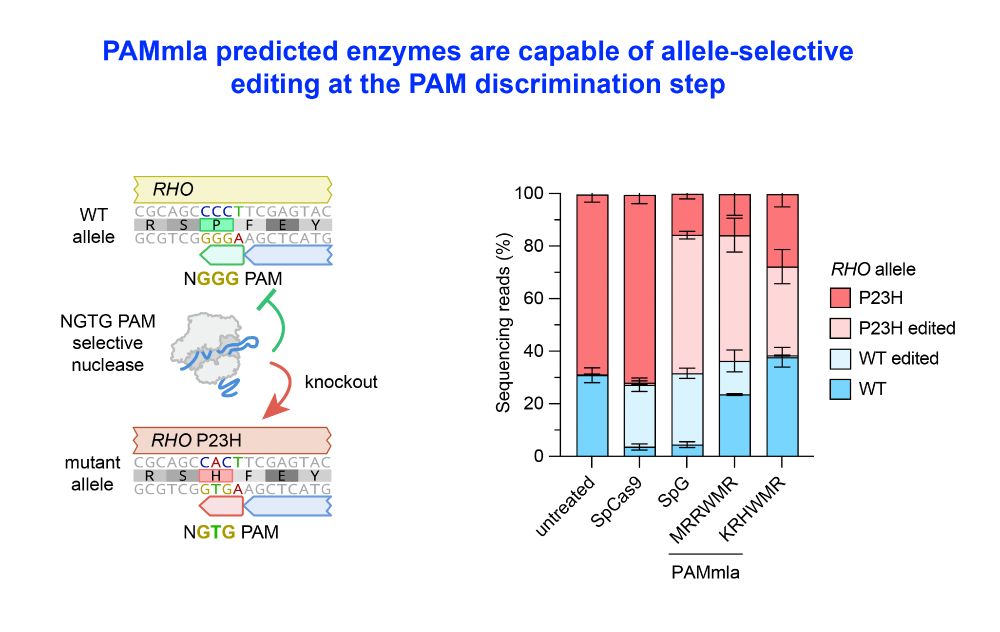
Importantly, PAMmla reversed the biology of Cas9, now able distinguish against a canonical NGGG PAM & instead effectively target an NGTG PAM.
pammla.streamlit.app
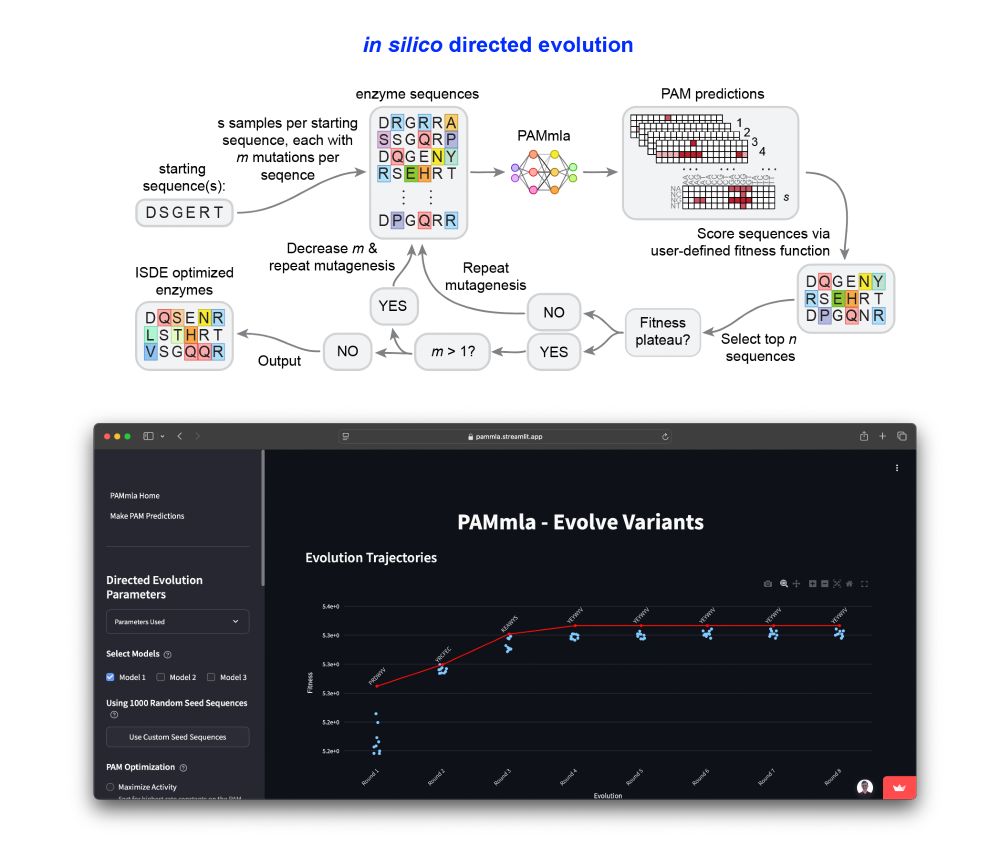
pammla.streamlit.app
No need to use PAM-relaxed enzymes like SpG anymore - use more specific PAMmla proteins instead!
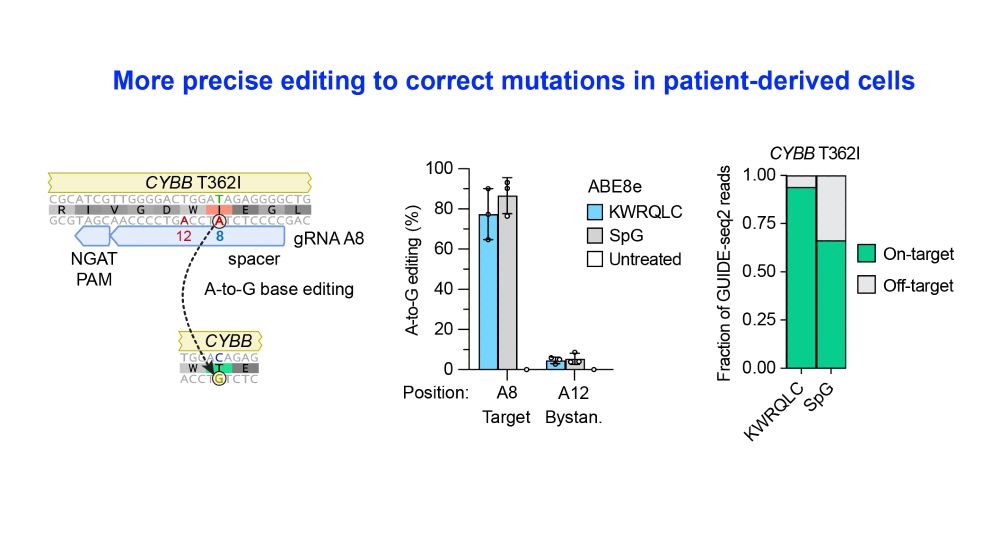
No need to use PAM-relaxed enzymes like SpG anymore - use more specific PAMmla proteins instead!
This results in safer enzymes that minimize unwanted off-target editing.
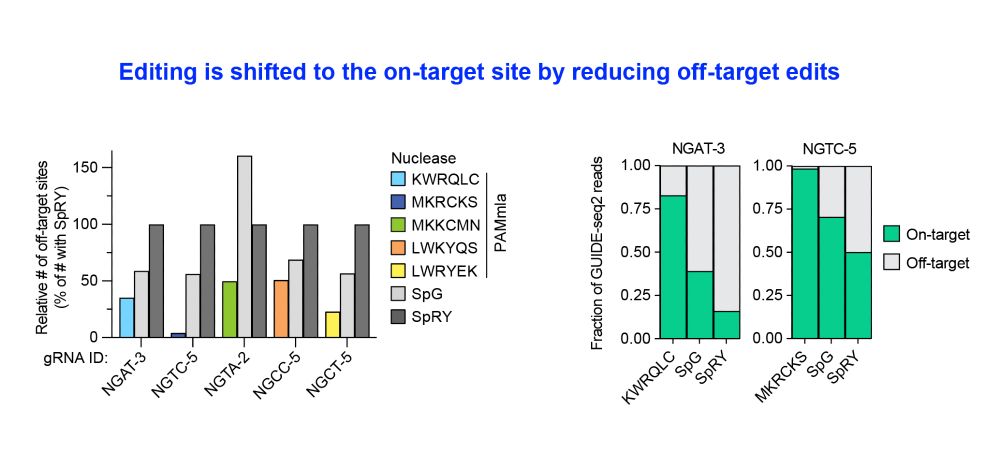
This results in safer enzymes that minimize unwanted off-target editing.
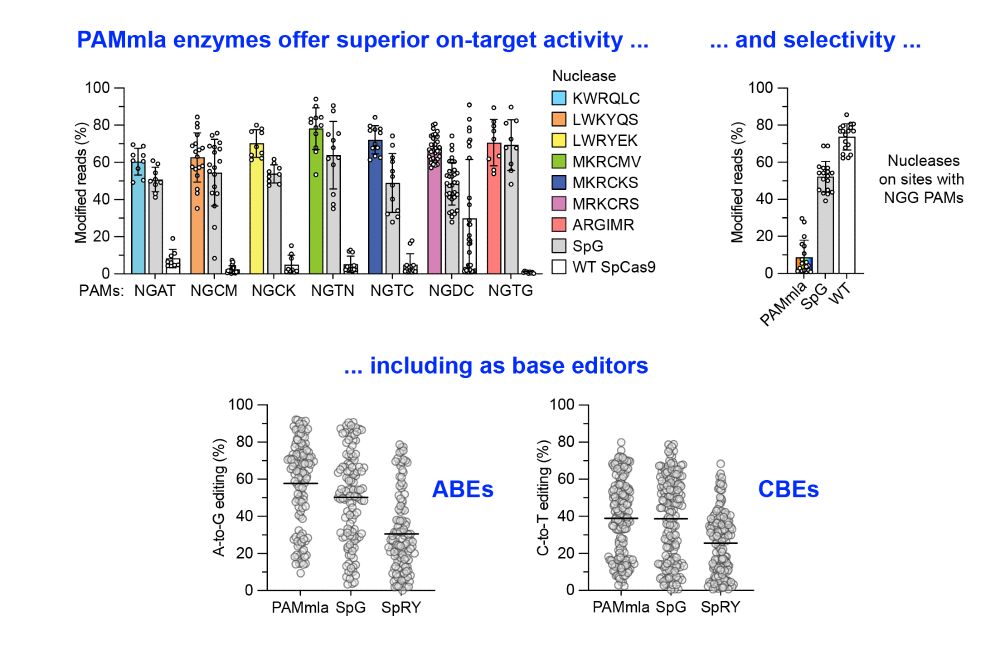
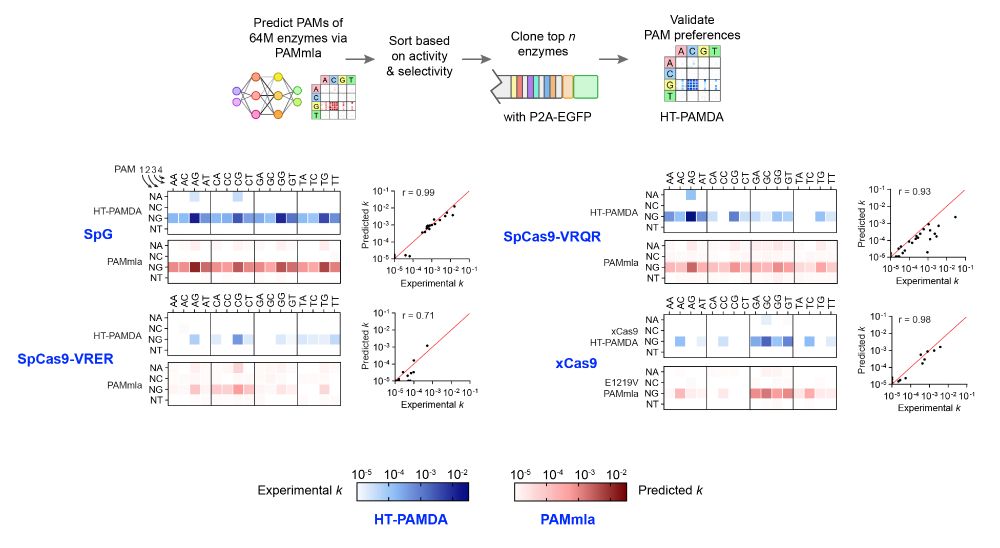
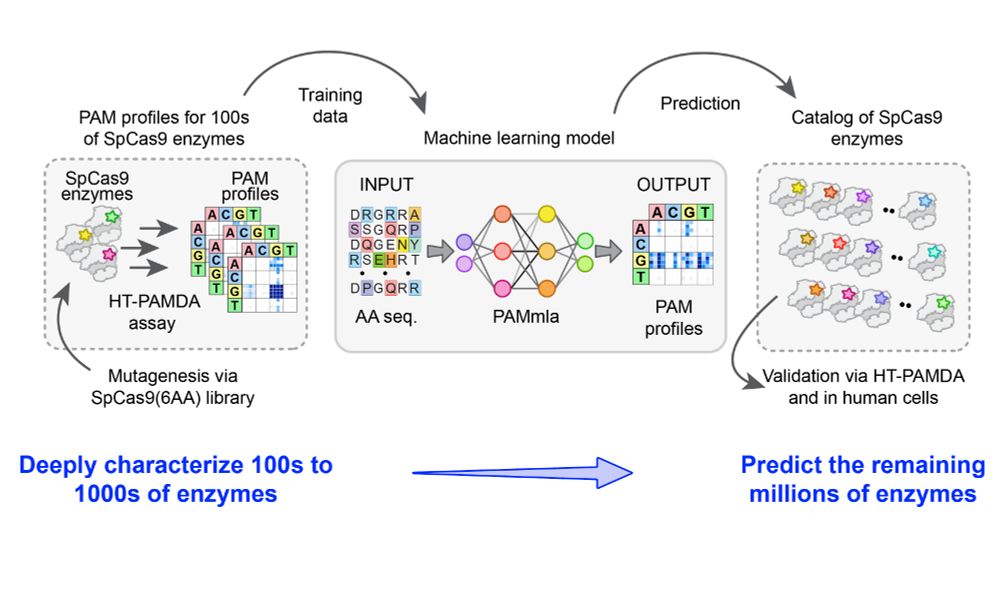
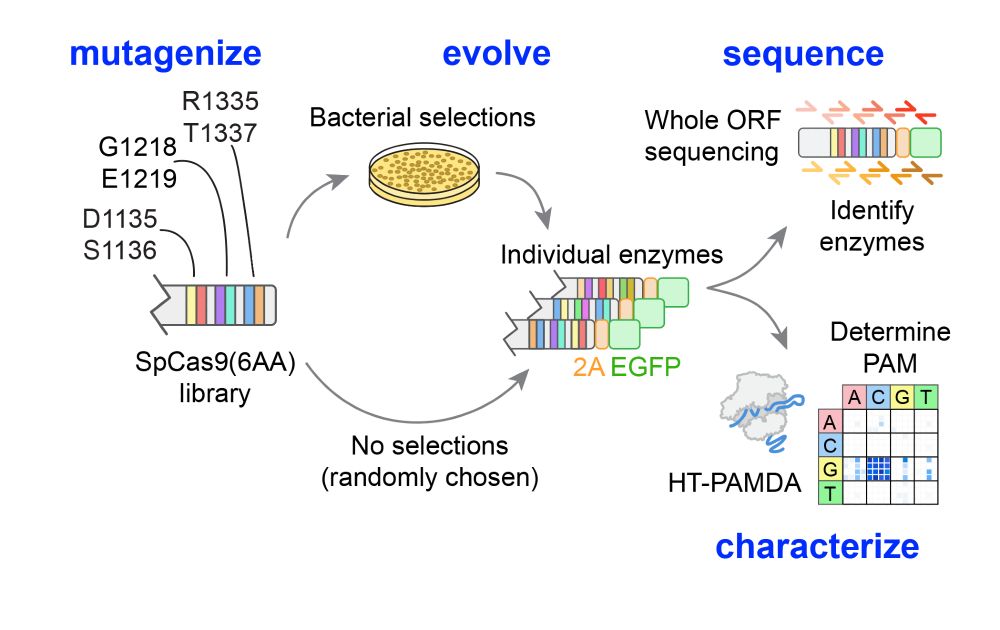

A massive collaborative effort including contributions from Nahye Kim @ann-sophiekroell.bsky.social @russelltwalton.bsky.social & many others.

A massive collaborative effort including contributions from Nahye Kim @ann-sophiekroell.bsky.social @russelltwalton.bsky.social & many others.


