Let’s talk about transparent, reproducible & frugal #AI for cancer research, the hallmarks of predictive oncology, and what’s next with agentic AI systems.
🔗 sites.google.com/ualberta.ca/...

Let’s talk about transparent, reproducible & frugal #AI for cancer research, the hallmarks of predictive oncology, and what’s next with agentic AI systems.
🔗 sites.google.com/ualberta.ca/...
Excited to work with exceptional colleagues across UHN to make this vision real!

Excited to work with exceptional colleagues across UHN to make this vision real!
Executive AI Scientific Director and Co-Director of the
@uhnaihub.bsky.social
This is a major opportunity to accelerate how we bring responsible, transparent and effective AI into patient care, research, education and operations across UHN.

Executive AI Scientific Director and Co-Director of the
@uhnaihub.bsky.social
This is a major opportunity to accelerate how we bring responsible, transparent and effective AI into patient care, research, education and operations across UHN.
Motion: Method developers should be required to publish all limitations & failures.
I’ll argue against mandatory transparency, not because I oppose open science, but to stress-test the idea and push our thinking.
🗓️ Nov 6, 11am EDT
🔗 rb.gy/p71tnv

Motion: Method developers should be required to publish all limitations & failures.
I’ll argue against mandatory transparency, not because I oppose open science, but to stress-test the idea and push our thinking.
🗓️ Nov 6, 11am EDT
🔗 rb.gy/p71tnv
The Data Science TeamLab is where AI and data science meet translational oncology, the sessions on (agentic) AI will chart new frontiers in how we cure cancer. 🚀
@break-cancer.bsky.social

The Data Science TeamLab is where AI and data science meet translational oncology, the sessions on (agentic) AI will chart new frontiers in how we cure cancer. 🚀
@break-cancer.bsky.social
We built TCL-38, the most comprehensive multi-omics + drug response atlas for TCL to predict selective drug sensitivities & combinations --> new therapeutic vulnerabilities & biomarkers.



We built TCL-38, the most comprehensive multi-omics + drug response atlas for TCL to predict selective drug sensitivities & combinations --> new therapeutic vulnerabilities & biomarkers.
Transparency, reproducibility & reusability aren’t optional—they’re the foundation for trustworthy AI in healthcare.
Join us: themaqc.org
#AI #MAQC25

Transparency, reproducibility & reusability aren’t optional—they’re the foundation for trustworthy AI in healthcare.
Join us: themaqc.org
#AI #MAQC25
🔍 Predicts novel ligand–receptor interactions
🧠 Handles spatial data & drug perturbations
🌐 Open-source: github.com/bhklab/GraphComm
Full paper: doi.org/10.1038/s415...


🔍 Predicts novel ligand–receptor interactions
🧠 Handles spatial data & drug perturbations
🌐 Open-source: github.com/bhklab/GraphComm
Full paper: doi.org/10.1038/s415...
New paper from @thesgc.bsky.social and led by UNC presents the first fully open AI framework for DNA-Encoded Library hit discovery, scanning billions of molecules and confirming novel binders.
👉 Data & models available on AIRCHECK aircheck.ai


New paper from @thesgc.bsky.social and led by UNC presents the first fully open AI framework for DNA-Encoded Library hit discovery, scanning billions of molecules and confirming novel binders.
👉 Data & models available on AIRCHECK aircheck.ai
Today marks our inaugural meeting, bringing together global experts to shape the future of AI-driven cancer research.

Today marks our inaugural meeting, bringing together global experts to shape the future of AI-driven cancer research.
I shared how AI and biomarker discovery are transforming precision oncology, and why rethinking therapy response evaluation is essential to deliver truly personalized cancer care.
Read more 👉 edition.pagesuite-professional.co.uk/html5/reader...


I shared how AI and biomarker discovery are transforming precision oncology, and why rethinking therapy response evaluation is essential to deliver truly personalized cancer care.
Read more 👉 edition.pagesuite-professional.co.uk/html5/reader...
Their method is called MorphDiff… which is super close to our MorphoDiff previously published at ICLR 2025.
Guess we’ll need a morphology diff checker now 😅
bsky.app/profile/bowa...
www.nature.com/articles/s41...


Their method is called MorphDiff… which is super close to our MorphoDiff previously published at ICLR 2025.
Guess we’ll need a morphology diff checker now 😅
bsky.app/profile/bowa...
www.nature.com/articles/s41...
So let me get this straight
1. Academics spend decades building the PDB openly
2. A company trains AlphaFold on it, releases limited code under pressure
3. Academics work hard to create fully open versions
4. Companies then use the open models to build closed products
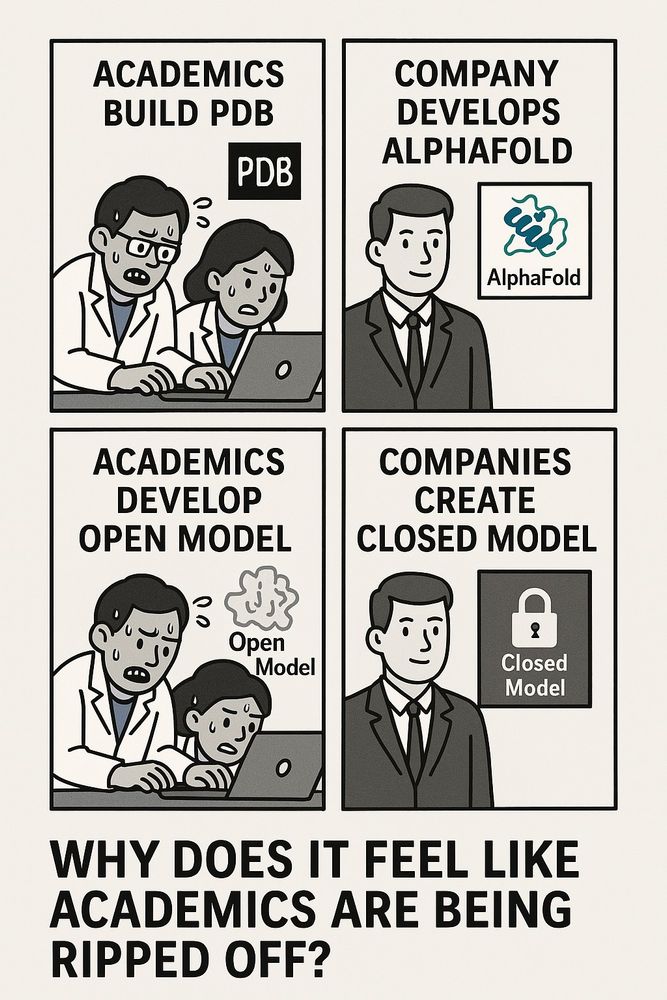
So let me get this straight
1. Academics spend decades building the PDB openly
2. A company trains AlphaFold on it, releases limited code under pressure
3. Academics work hard to create fully open versions
4. Companies then use the open models to build closed products
Here are the slides: bit.ly/3S9D6b1
They are open-source so feel free to re-use.
Any feedback is welcome!

Here are the slides: bit.ly/3S9D6b1
They are open-source so feel free to re-use.
Any feedback is welcome!
bit.ly/47iIRsq

bit.ly/47iIRsq
The package is based on our recent paper in Annals of Oncology:
doi.org/10.1016/j.an...

The package is based on our recent paper in Annals of Oncology:
doi.org/10.1016/j.an...

