What other methods did you run into? Did you plan on using another medium?

What other methods did you run into? Did you plan on using another medium?
Find this option in the "Linear Synteny" or "Dotplot" alignments views (bottom), or in separate data tracks (per genome).
Thanks for the excellent reviewer comments on this.
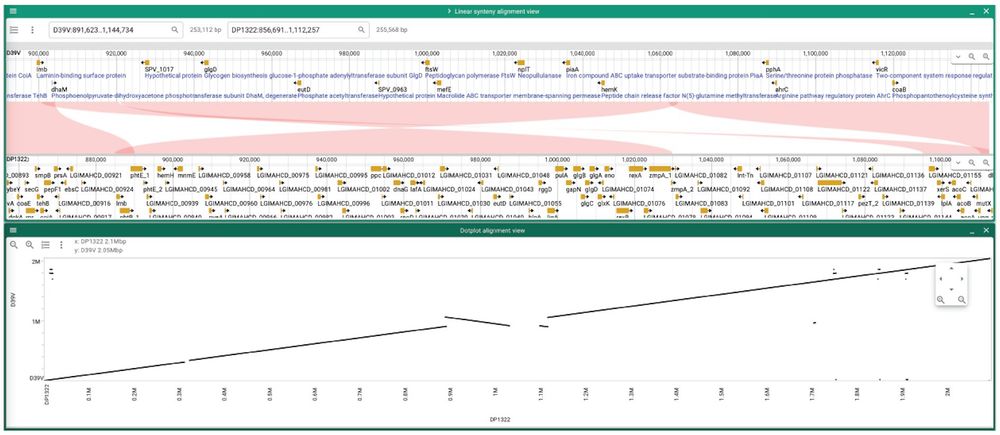
Find this option in the "Linear Synteny" or "Dotplot" alignments views (bottom), or in separate data tracks (per genome).
Thanks for the excellent reviewer comments on this.
We hope you enjoy!

We hope you enjoy!