
Postdoc in the Whitehouse lab, MSKCC, NYC.
Former PhD in the Pasero lab, IGH, France.
10/11

10/11
Not all fuzziness is due to transcription.
Rather, the genomic distance between two consecutive NFRs (Nuc Free Region) predict fuzziness. If the size is a multiple of the optimal Nucleosome Repeat Length (NRL) => Positioned Array. If not => Delocalized Array.
9/11

Not all fuzziness is due to transcription.
Rather, the genomic distance between two consecutive NFRs (Nuc Free Region) predict fuzziness. If the size is a multiple of the optimal Nucleosome Repeat Length (NRL) => Positioned Array. If not => Delocalized Array.
9/11
8/11

8/11
7/11

7/11
6/11

6/11
5/11

5/11
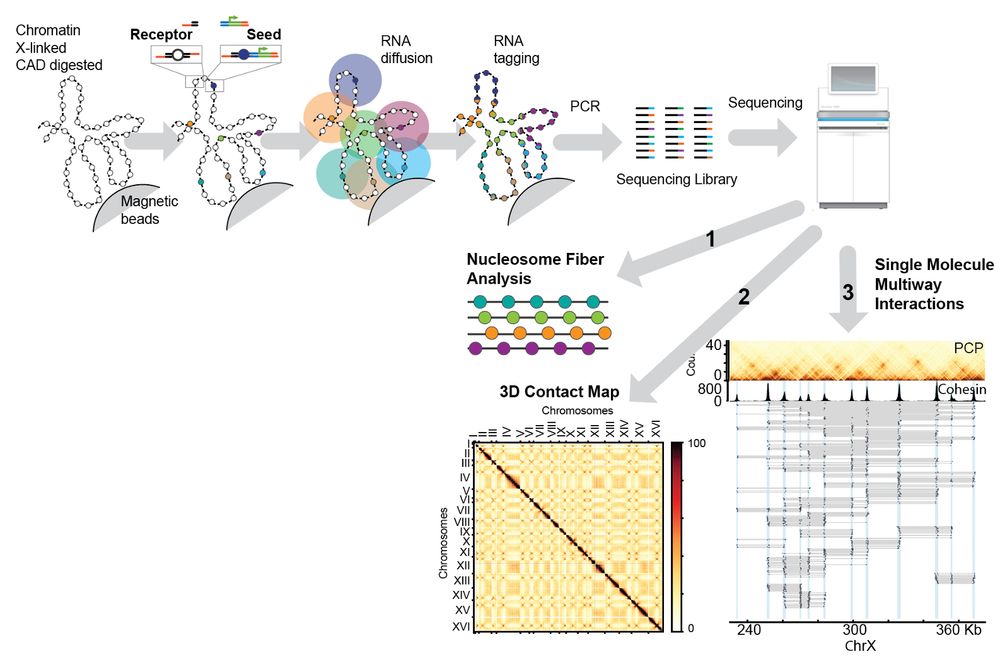
Chromatin is X-linked and digested to mononucs, a mixture of Seeds and Receptors is ligated to DNA ends. Each seed will produce unique tagRNAs and tag nucs in proximity.
After sequencing, reads are grouped based on the UMI, informing on spatial proximity.

Chromatin is X-linked and digested to mononucs, a mixture of Seeds and Receptors is ligated to DNA ends. Each seed will produce unique tagRNAs and tag nucs in proximity.
After sequencing, reads are grouped based on the UMI, informing on spatial proximity.
• A Seed is transcribed to produce a tagRNA carrying: a unique identifier (UMI) and an annealing sequence.
• The tagRNAs diffuse and anneal to Receptors sharing the annealing sequence
• The annealed tagRNA is copied by RT to “tag” the Receptor
3/11

• A Seed is transcribed to produce a tagRNA carrying: a unique identifier (UMI) and an annealing sequence.
• The tagRNAs diffuse and anneal to Receptors sharing the annealing sequence
• The annealed tagRNA is copied by RT to “tag” the Receptor
3/11
The latest method from the Whitehouse lab: PCP
PCP uses a novel proximity barcoding strategy to simultaneously map 3D genome organization at various resolutions at the single molecule level:
Out soon, check the updated BioRxiv – little thread:
1/11
www.biorxiv.org/content/10.1...
The latest method from the Whitehouse lab: PCP
PCP uses a novel proximity barcoding strategy to simultaneously map 3D genome organization at various resolutions at the single molecule level:
Out soon, check the updated BioRxiv – little thread:
1/11
www.biorxiv.org/content/10.1...
Can follow on zoom here: nyulangone.zoom.us/webinar/regi...

Can follow on zoom here: nyulangone.zoom.us/webinar/regi...

