Evolution, immunity, genomics, microbiolgy.
Into immunity in bacteria and its conservation in eukaryotes.
Advocate for more inclusive sciences
https://research.pasteur.fr/en/team/molecular-diversity-of-microbes/
SIRal, a human homolog, is a novel actor of the TLR pathway.
Much remains to be understood about SIRal and Sirims in immunity, but 🦠 will help us along the way.
SIRal, a human homolog, is a novel actor of the TLR pathway.
Much remains to be understood about SIRal and Sirims in immunity, but 🦠 will help us along the way.
Like their bacterial relatives, eukaryotic SIRims — including human SIRal — use NAD+ in vitro. This enzymatic activity (NADase) seems ancient and conserved across the tree of life.
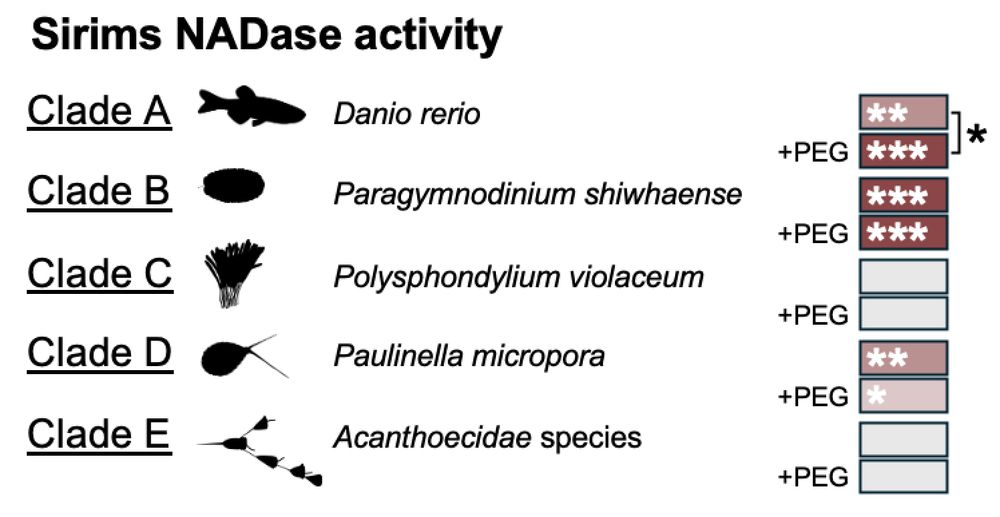
Like their bacterial relatives, eukaryotic SIRims — including human SIRal — use NAD+ in vitro. This enzymatic activity (NADase) seems ancient and conserved across the tree of life.
We demonstrate that SIRal limits infection by herpes virus and by the bacteria Salmonella in mouse macrophages.

We demonstrate that SIRal limits infection by herpes virus and by the bacteria Salmonella in mouse macrophages.
Through numerous experiments (yay bar graphs!), we show that SIRal is required for a key immune pathway in animals (Toll-like receptor/TLR). SIRal mediates the up-regulation of interferon-stimulated genes (ISGs) and proinflammatory cytokines.

Through numerous experiments (yay bar graphs!), we show that SIRal is required for a key immune pathway in animals (Toll-like receptor/TLR). SIRal mediates the up-regulation of interferon-stimulated genes (ISGs) and proinflammatory cytokines.
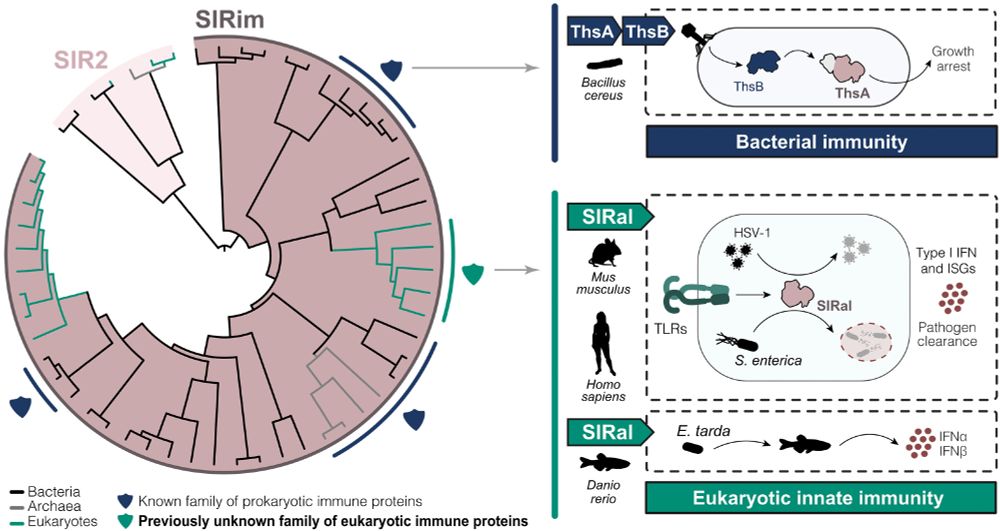
Among this large family of proteins, present in thousands of living organisms, we identified on human protein, which function was previously unknown.
We named it SIRal, for SIR2 antiphage-like.

Among this large family of proteins, present in thousands of living organisms, we identified on human protein, which function was previously unknown.
We named it SIRal, for SIR2 antiphage-like.
We analyzed SIR2 from thousands of bacterial, archaeal & eukaryotic genomes. All housekeeping sirtuins group in a clade, whereas all antiphage SIR2 group in another one that we named SIRim, for “SIR” and “immunity”.

We analyzed SIR2 from thousands of bacterial, archaeal & eukaryotic genomes. All housekeeping sirtuins group in a clade, whereas all antiphage SIR2 group in another one that we named SIRim, for “SIR” and “immunity”.
Check out their work 👇


Check out their work 👇
Really fun to be celebrating the contribution of diverse organisms to our understanding of immunity 🪰🐘🦠.

Really fun to be celebrating the contribution of diverse organisms to our understanding of immunity 🪰🐘🦠.
Vive la science, et vive Paris!




Vive la science, et vive Paris!
So excited for three days of awesome science.




So excited for three days of awesome science.
We built an interactive UMAP to geek this space mdmparis.github.io/antiphage-la...
Quantitatively, rarefaction curves led us to estimate that probably >45k prot families are antiphage

We built an interactive UMAP to geek this space mdmparis.github.io/antiphage-la...
Quantitatively, rarefaction curves led us to estimate that probably >45k prot families are antiphage
Geb is only 150AA protein with no annotated Pfam domain while Ukko is a 4 genes system with intriguing links to metabolism.
Going beyond classic model organisms is awesome for novel biology.

Geb is only 150AA protein with no annotated Pfam domain while Ukko is a 4 genes system with intriguing links to metabolism.
Going beyond classic model organisms is awesome for novel biology.
By novel, we mean 1)not picked up by defense score, and 2) with proteins with domains never involved in antiphage defense before.
We validated six novel antiphage systems in Streptomyces (out of 10 tested)!

By novel, we mean 1)not picked up by defense score, and 2) with proteins with domains never involved in antiphage defense before.
We validated six novel antiphage systems in Streptomyces (out of 10 tested)!
Some systems found in hotspots of MGE could be missed by defense score, and some novel systems without known antiphage domains by ESM.

Some systems found in hotspots of MGE could be missed by defense score, and some novel systems without known antiphage domains by ESM.
We focused on Actinobacteria that have been underexplored for their antiphage repertoire.
👋ALBERT-DefenseFinder🧐

We focused on Actinobacteria that have been underexplored for their antiphage repertoire.
👋ALBERT-DefenseFinder🧐
We show ESM-DefenseFinder can predict many novel antiphage proteins, which represent variants of known systems as it relies mostly on homology.

We show ESM-DefenseFinder can predict many novel antiphage proteins, which represent variants of known systems as it relies mostly on homology.
But computational approaches like defense islands, MGE hotspots suggest thousands remain undescribed. How big is this unknown?

But computational approaches like defense islands, MGE hotspots suggest thousands remain undescribed. How big is this unknown?
We used protein and genomic language models (and defense score!) to start bringing some answers: >45 000 protein families.
Leb by E. Mordret.
www.biorxiv.org/content/10.1...
Interactive UMAP to have fun
mdmparis.github.io/antiphage-la...
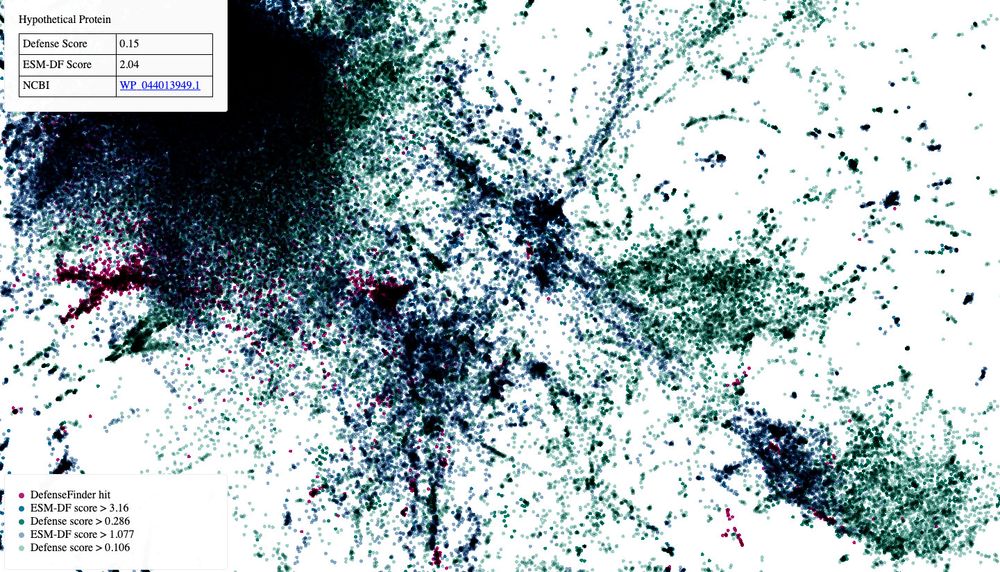
We used protein and genomic language models (and defense score!) to start bringing some answers: >45 000 protein families.
Leb by E. Mordret.
www.biorxiv.org/content/10.1...
Interactive UMAP to have fun
mdmparis.github.io/antiphage-la...
www.sisb2025.conferences-pasteur.org/home
Joins us in Institut Pasteur, Paris, April 8-10th.

www.sisb2025.conferences-pasteur.org/home
Joins us in Institut Pasteur, Paris, April 8-10th.
Such a beautiful place, moved and happy ++ to share it with Hugo, Florian and Jean who started the lab with me, and in presence of scientists who inspire me !




Such a beautiful place, moved and happy ++ to share it with Hugo, Florian and Jean who started the lab with me, and in presence of scientists who inspire me !
Excited to welcome these amazing speakers at Institut Pasteur.
Registration will open beginning of November. Abstract deadline February 28. Stay tuned.
Co-organized by @soreklab.bsky.social P. Kranzusch

Excited to welcome these amazing speakers at Institut Pasteur.
Registration will open beginning of November. Abstract deadline February 28. Stay tuned.
Co-organized by @soreklab.bsky.social P. Kranzusch
In total 19% of queried eukaryotic genomes encode a sirim, spanning 189 very diverse species. So much to explore 🤩!

In total 19% of queried eukaryotic genomes encode a sirim, spanning 189 very diverse species. So much to explore 🤩!
Bacterial SIRim degrade NAD+, could SIRanc also degrade NAD+? Indeed! SIRanc has NADase activity in vitro, and this activity is necessary for SIRanc in vivo.
So SIRanc like bacterial SIRim is an NADase!

Bacterial SIRim degrade NAD+, could SIRanc also degrade NAD+? Indeed! SIRanc has NADase activity in vitro, and this activity is necessary for SIRanc in vivo.
So SIRanc like bacterial SIRim is an NADase!
=> SIRanc contributes to TLR signalling!

=> SIRanc contributes to TLR signalling!
Through a series of experiments, we show that SIRanc is involved in the TRIF dependent (and not MyD88) branch of the TLR4 pathway in mice, a reason why it could have been missed by other screens !
But what about humans?

Through a series of experiments, we show that SIRanc is involved in the TRIF dependent (and not MyD88) branch of the TLR4 pathway in mice, a reason why it could have been missed by other screens !
But what about humans?

