
IMO, data converges: we need to revisit the atlas.
But I agree broadly: brain tissue is far from homogenous, and we neglect it at our peril.
IMO, data converges: we need to revisit the atlas.
But I agree broadly: brain tissue is far from homogenous, and we neglect it at our peril.
Yet if there is controversy around how areas are defined from neural recordings, cell diversity is equally as confusing a signal.... (1/2)

Yet if there is controversy around how areas are defined from neural recordings, cell diversity is equally as confusing a signal.... (1/2)
If you can't quantify natural language uncertainty, you can't train for it. At least explicitly. Thus all we have are RLHF approaches
If you can't quantify natural language uncertainty, you can't train for it. At least explicitly. Thus all we have are RLHF approaches
If you gave me this measure for all data in the wild, and it was differentiable wrt the model, one might take a likelihood-max approach (plus calibration etc)
If you gave me this measure for all data in the wild, and it was differentiable wrt the model, one might take a likelihood-max approach (plus calibration etc)
1) gets the avg (pseudobulk) gene expression of each type,
2) computes the similarity matrix,
3) projects expression into a 3D space using MDS, and
4) interprets the space as a perceptually uniform color space (LUV)
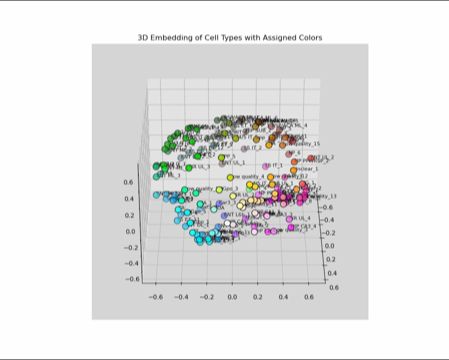
1) gets the avg (pseudobulk) gene expression of each type,
2) computes the similarity matrix,
3) projects expression into a 3D space using MDS, and
4) interprets the space as a perceptually uniform color space (LUV)
```
!pip install colormycells
# Create a colormap based on cell type similarities
colors = get_colormap(adata, key="cell_type")
# Plot your cells
sc.pl.umap(adata, color="cell_type", palette=colors)
````
github.com/ZadorLaborat...
```
!pip install colormycells
# Create a colormap based on cell type similarities
colors = get_colormap(adata, key="cell_type")
# Plot your cells
sc.pl.umap(adata, color="cell_type", palette=colors)
````
github.com/ZadorLaborat...
I also think this makes pretty plots 🌈 so that's nice.
I also think this makes pretty plots 🌈 so that's nice.
(-) some cell types jump out for no reason
(-) perceptual similarity between colors is meaningless
(-) fewer colors than types, leading to repeats

(-) some cell types jump out for no reason
(-) perceptual similarity between colors is meaningless
(-) fewer colors than types, leading to repeats
Rather than the bell tolling for local specialization, it might be tolling for that ‘functional’ stim-response paradigm
Rather than the bell tolling for local specialization, it might be tolling for that ‘functional’ stim-response paradigm


