
Brett Baker
@archaeal.bsky.social
Professor of microbial ecology and evolution at Univ of Texas Austin.
Baker Lab 2025
The container is our field gear for the research expedition for Uruguay.
sites.utexas.edu/baker-lab/sp...
The container is our field gear for the research expedition for Uruguay.
sites.utexas.edu/baker-lab/sp...
September 18, 2025 at 9:35 PM
Baker Lab 2025
The container is our field gear for the research expedition for Uruguay.
sites.utexas.edu/baker-lab/sp...
The container is our field gear for the research expedition for Uruguay.
sites.utexas.edu/baker-lab/sp...
Title: The phylogeny and metabolism underlying the global dominance of the freshwater Nanopelagicaceae lineage
Short Talk Symposia: Hot Topics in Microbial Diversity and Systematics
Session 6/21/2025 1:45- 3:45:00 403A
Presentation Time: 3:15:00 PM
Short Talk Symposia: Hot Topics in Microbial Diversity and Systematics
Session 6/21/2025 1:45- 3:45:00 403A
Presentation Time: 3:15:00 PM

June 21, 2025 at 2:25 PM
Title: The phylogeny and metabolism underlying the global dominance of the freshwater Nanopelagicaceae lineage
Short Talk Symposia: Hot Topics in Microbial Diversity and Systematics
Session 6/21/2025 1:45- 3:45:00 403A
Presentation Time: 3:15:00 PM
Short Talk Symposia: Hot Topics in Microbial Diversity and Systematics
Session 6/21/2025 1:45- 3:45:00 403A
Presentation Time: 3:15:00 PM
Tomorrow at 12:45-1:30 in the EEB Track Hub in the poster hall #ASMmicrobe I am talking about microbial diversity including Asgard archaea, and an expanded tree of life from marine sediments.

June 20, 2025 at 3:23 PM
Tomorrow at 12:45-1:30 in the EEB Track Hub in the poster hall #ASMmicrobe I am talking about microbial diversity including Asgard archaea, and an expanded tree of life from marine sediments.
The Baker Lab is invading LA for #ASMMicrobe


June 19, 2025 at 1:16 PM
The Baker Lab is invading LA for #ASMMicrobe
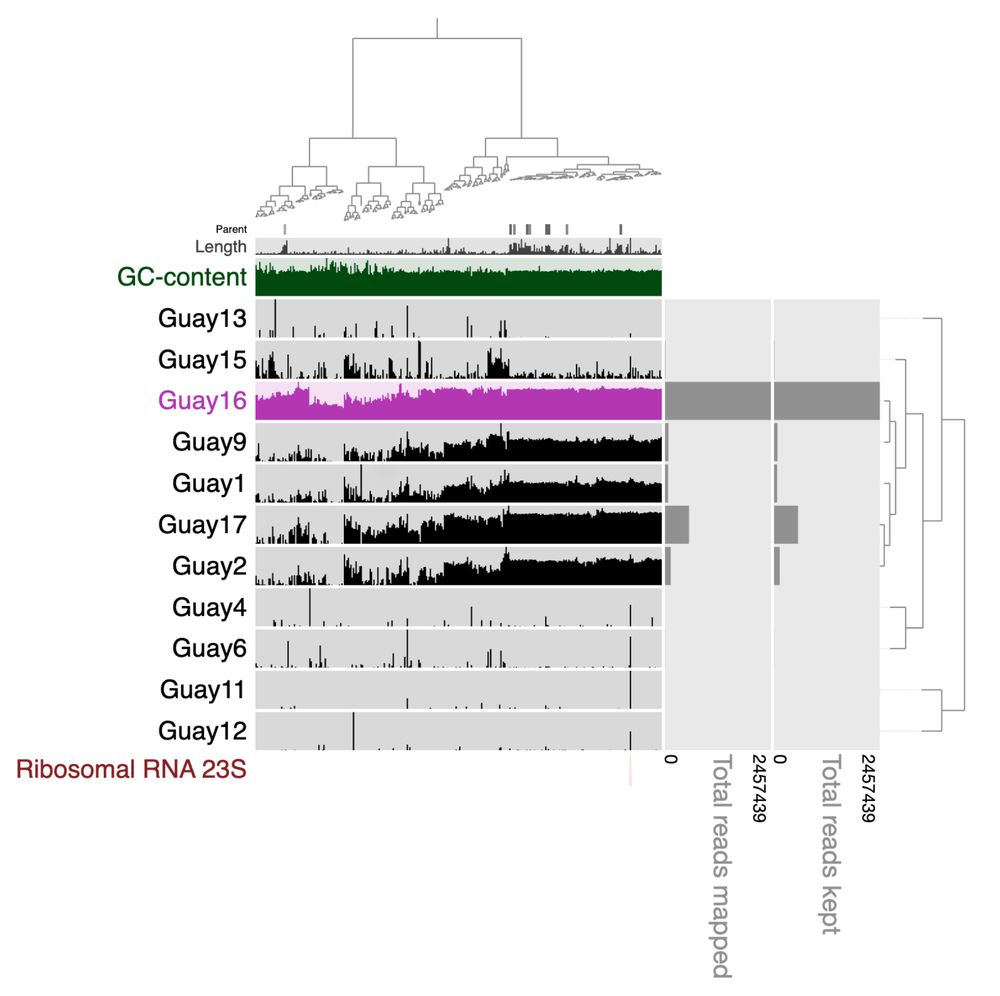
Well said, but not well represented in the paper. Another example, B6, they claim to be contaminated (shown in Fig 2a). Green are the correct reads from which the MAG was generated. Location markers used for phylogeny in Eme et al in pink.

May 15, 2025 at 2:34 PM
Well said, but not well represented in the paper. Another example, B6, they claim to be contaminated (shown in Fig 2a). Green are the correct reads from which the MAG was generated. Location markers used for phylogeny in Eme et al in pink.
Pink is the sample the MAG came from! Are there misbinned contigs, perhaps, we are checking now… However, Njordarchaeales are commonly removed from phylogenies due to their unique nature.
May 8, 2025 at 12:23 PM
Pink is the sample the MAG came from! Are there misbinned contigs, perhaps, we are checking now… However, Njordarchaeales are commonly removed from phylogenies due to their unique nature.
The cyclases in this pathways are ancestral to a dipternoind, present in modern plants! Heterologous expression of them revealed they cyclize geranylgeranyl pyrophosphate to form bicyclic halimadienyl pyrophosphate.

February 8, 2025 at 9:35 PM
The cyclases in this pathways are ancestral to a dipternoind, present in modern plants! Heterologous expression of them revealed they cyclize geranylgeranyl pyrophosphate to form bicyclic halimadienyl pyrophosphate.
Eukaryotic membranes have polycyclic triterpenoids (mainly sterols) that are essential for a variety of cellular functions, but these have not been seen in archaea. So we searched for them in Asgard archaea, which are the closest relative to eukaryotes. This revealed biosynthetic pathways for them.

February 8, 2025 at 9:32 PM
Eukaryotic membranes have polycyclic triterpenoids (mainly sterols) that are essential for a variety of cellular functions, but these have not been seen in archaea. So we searched for them in Asgard archaea, which are the closest relative to eukaryotes. This revealed biosynthetic pathways for them.
In an accompanying study we tracked the viral ecology of the same samples in collaboration with @karthik-a.bsky.social team at UW t.co/nj4QSA7nXE

January 3, 2025 at 12:07 PM
In an accompanying study we tracked the viral ecology of the same samples in collaboration with @karthik-a.bsky.social team at UW t.co/nj4QSA7nXE
We looked at specific genes under selection and found that in a dominant bacterium, there was a dramatic shift in this population in 2012, in which genes under selection are involved in the degradation of biomolecules. This coincides with environmental extremes that summer.

January 3, 2025 at 12:05 PM
We looked at specific genes under selection and found that in a dominant bacterium, there was a dramatic shift in this population in 2012, in which genes under selection are involved in the degradation of biomolecules. This coincides with environmental extremes that summer.
Given the long-term data, we also examine decadal shift in strain composition and found that dominant bacterial strains experience gradual change, others have step type shifts, as well as disturbance and resilience.

January 3, 2025 at 12:02 PM
Given the long-term data, we also examine decadal shift in strain composition and found that dominant bacterial strains experience gradual change, others have step type shifts, as well as disturbance and resilience.
We leverage this astounding dataset to track both community structure (ecology) and evolution at the strain level. Of course, abundance of dominant bacteria is season, but interestingly so does single nucleotide variants.

January 3, 2025 at 12:00 PM
We leverage this astounding dataset to track both community structure (ecology) and evolution at the strain level. Of course, abundance of dominant bacteria is season, but interestingly so does single nucleotide variants.
These metagenomes were used to constructed over 85,000 MAGs! Which were reduced to 2,855 representative genotypes. The dominant taxa belong to Nanopelagicales, Actinotbacteria. We used this amazing dataset this to look at long term ecology and evolution.

January 3, 2025 at 11:58 AM
These metagenomes were used to constructed over 85,000 MAGs! Which were reduced to 2,855 representative genotypes. The dominant taxa belong to Nanopelagicales, Actinotbacteria. We used this amazing dataset this to look at long term ecology and evolution.
New preprint, Methane production from hydrocarbon by consortium dominated by ANME archaea.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


October 31, 2024 at 1:37 PM
New preprint, Methane production from hydrocarbon by consortium dominated by ANME archaea.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
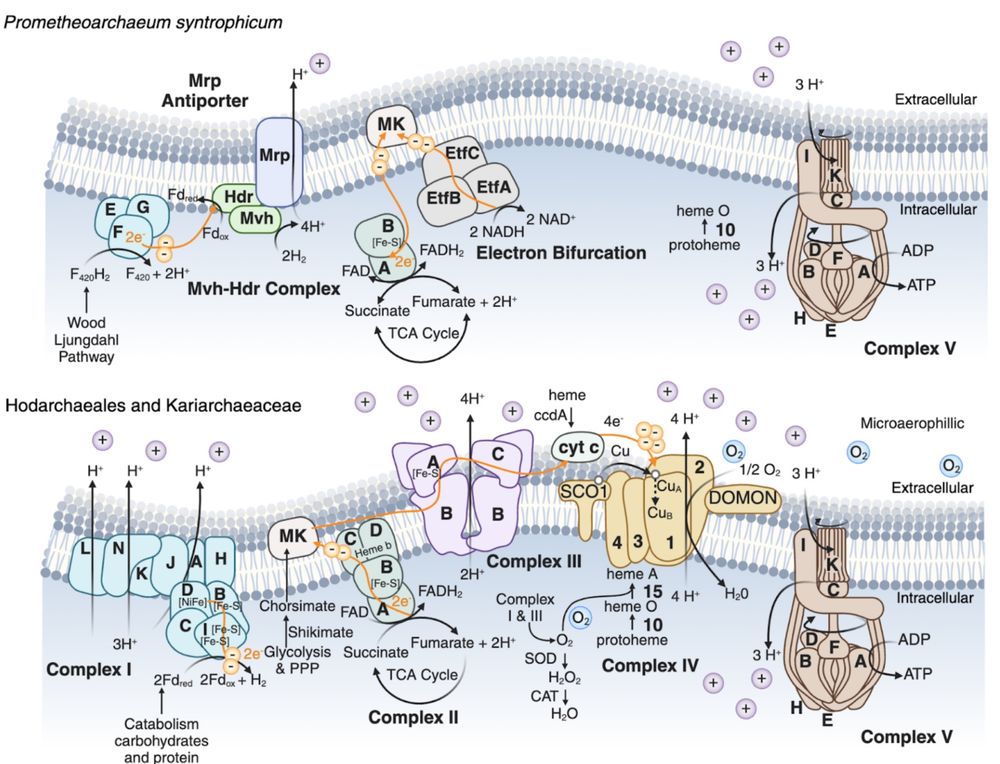
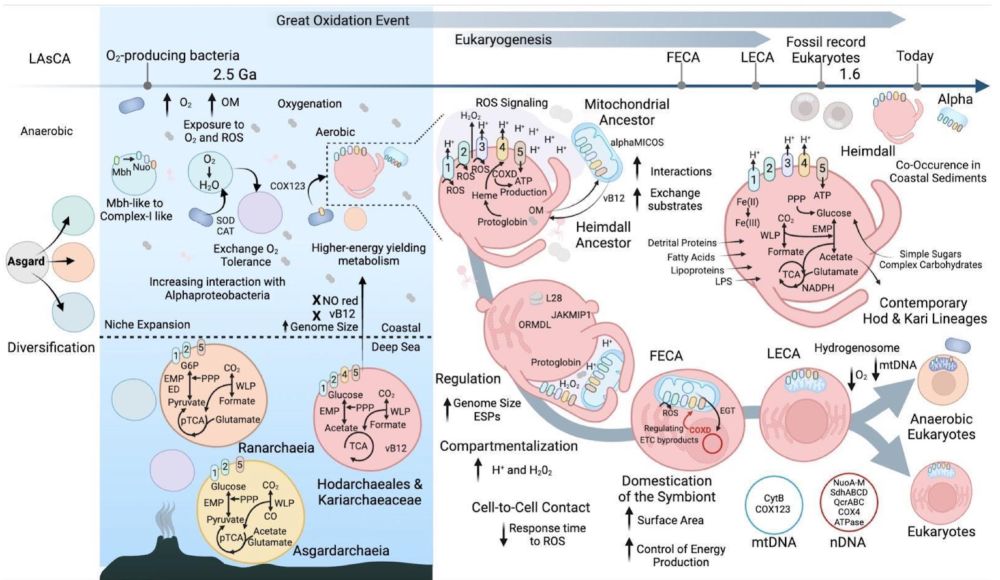
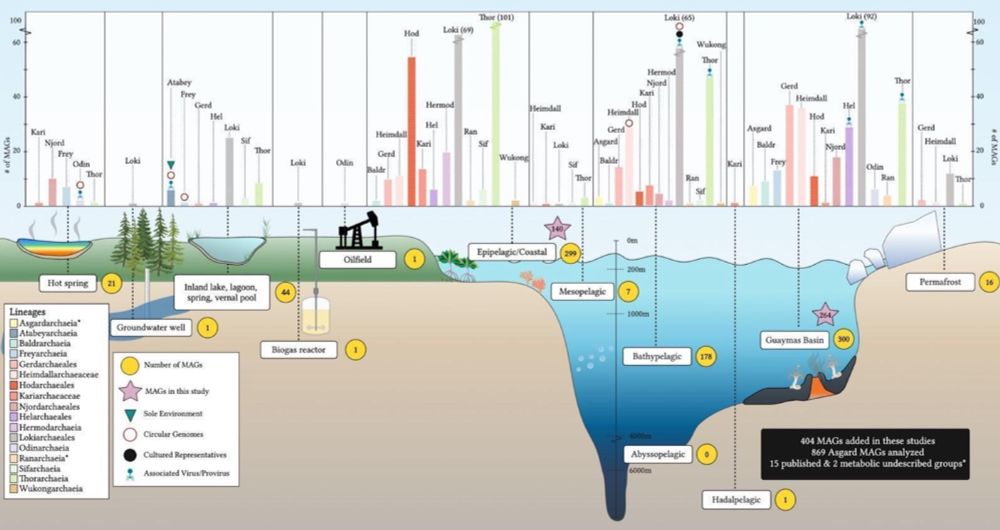
After many years of work we present a preprint that doubles the numbers of Asgard genomes, revealing lots of new insights into their metabolism. Notably, lineages closely related to eukaryotes are capable of aerobic respiration!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
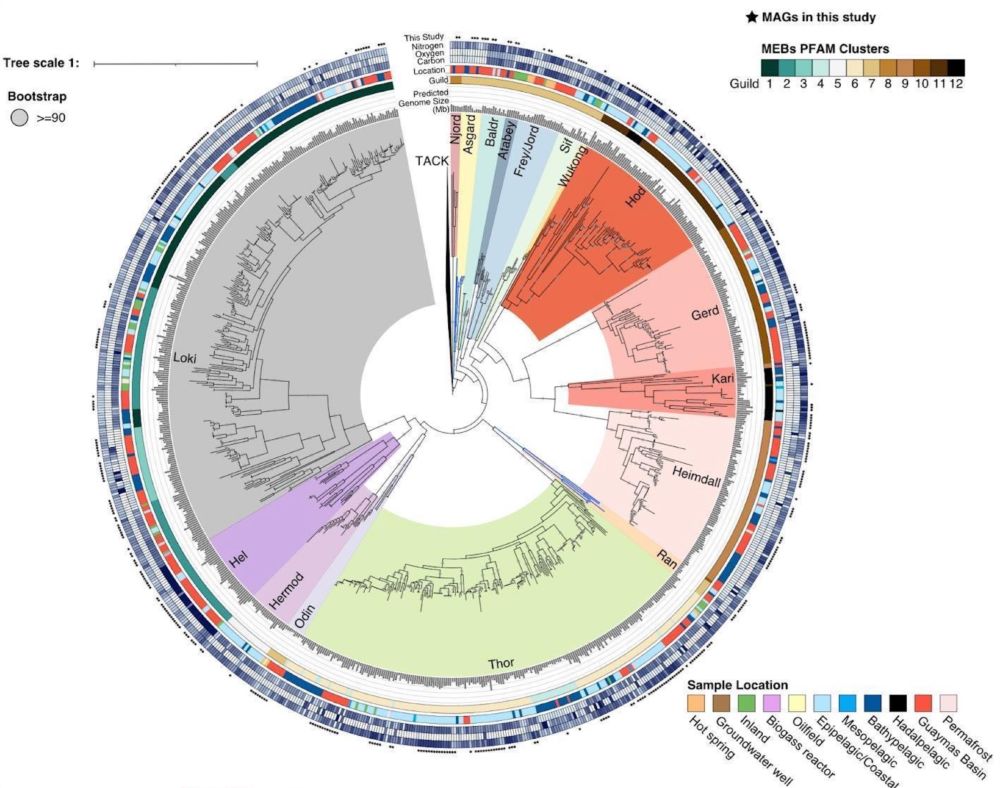
July 5, 2024 at 1:51 PM
After many years of work we present a preprint that doubles the numbers of Asgard genomes, revealing lots of new insights into their metabolism. Notably, lineages closely related to eukaryotes are capable of aerobic respiration!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Also see the accompanying study, that tracks viruses over all 20 years!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...




February 8, 2024 at 4:51 PM
Also see the accompanying study, that tracks viruses over all 20 years!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Very excited to announce a 20 year study of microbial ecology and evolution in a lake out today. This is the largest metagenomic seqing of a site resulting in >30k MAGs. We track shift in diversity and strains, and link sweeps to environmental factors.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...




February 8, 2024 at 4:39 PM
Very excited to announce a 20 year study of microbial ecology and evolution in a lake out today. This is the largest metagenomic seqing of a site resulting in >30k MAGs. We track shift in diversity and strains, and link sweeps to environmental factors.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Our latest preprint - Globally distributed marine Gemmatimonadota have unique genomic potentials
www.researchsquare.com/article/rs-3...
www.researchsquare.com/article/rs-3...


January 25, 2024 at 4:03 AM
Our latest preprint - Globally distributed marine Gemmatimonadota have unique genomic potentials
www.researchsquare.com/article/rs-3...
www.researchsquare.com/article/rs-3...
Our new publication looking at novel bacteria physiologies of alkane degradation in the North Atlantic.
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...


September 13, 2023 at 3:37 PM
Our new publication looking at novel bacteria physiologies of alkane degradation in the North Atlantic.
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...






