Can't possibly be happier to be working on phage satellites with @jmouradesousa.bsky.social , @epcrocha.bsky.social and the team!

Can't possibly be happier to be working on phage satellites with @jmouradesousa.bsky.social , @epcrocha.bsky.social and the team!
An important chapter of my life and career comes to an end - now I'm ready and excited for the next!




An important chapter of my life and career comes to an end - now I'm ready and excited for the next!





mad1 as putative system regulator
mad2:mad5 to form the N6 Meth complex
mad3-4 predicted nucleases
mad6 could regulate the system via phosphorylation
MAD7 may help recognize unmethylated DNA for loading into the MAD8 nuclease

mad1 as putative system regulator
mad2:mad5 to form the N6 Meth complex
mad3-4 predicted nucleases
mad6 could regulate the system via phosphorylation
MAD7 may help recognize unmethylated DNA for loading into the MAD8 nuclease
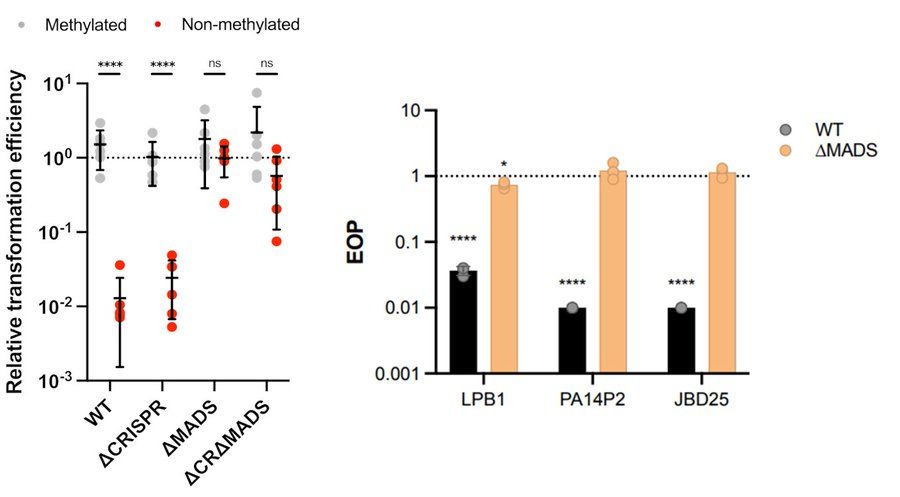
Single resistance: phages can easily bypass each defense
Dual resistance: Acr-phages must first acquire methylation to bypass MADS to successfully cooperate to overcome CRISPR, but this process is constrained by CRISPR activity

Single resistance: phages can easily bypass each defense
Dual resistance: Acr-phages must first acquire methylation to bypass MADS to successfully cooperate to overcome CRISPR, but this process is constrained by CRISPR activity
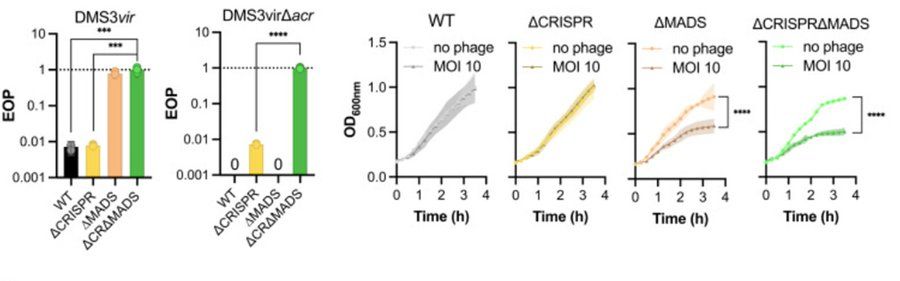
Such high starting phage density is not required to support phage persistence when CRISPR-Cas is absent (yellow)

Such high starting phage density is not required to support phage persistence when CRISPR-Cas is absent (yellow)



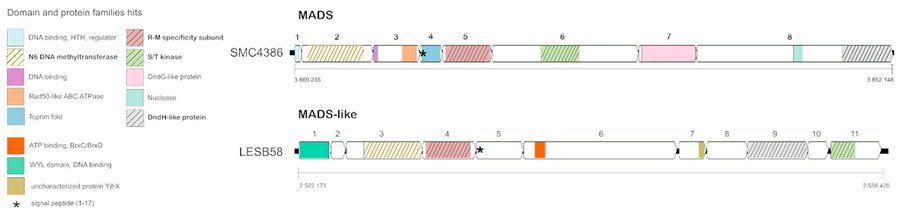
It is located within a genomic hotspot with conserved boundaries
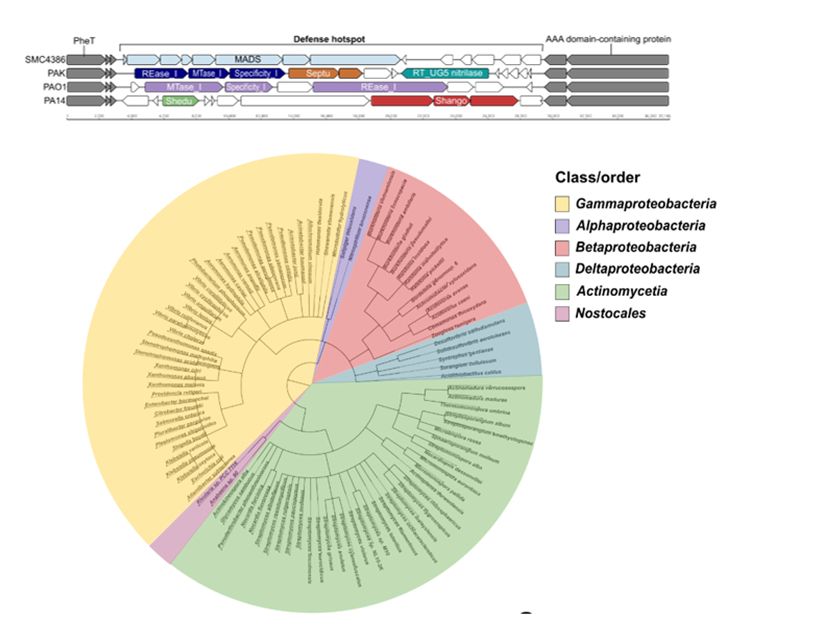
It is located within a genomic hotspot with conserved boundaries
These include:
- N6 methyltransferase domain
- serine/threonine kinase domain
- RM specificity subunit
- Dnd-like proteins
- DNA binding and nuclease domains

These include:
- N6 methyltransferase domain
- serine/threonine kinase domain
- RM specificity subunit
- Dnd-like proteins
- DNA binding and nuclease domains
so we hypothesized another system may be involved 👀

so we hypothesized another system may be involved 👀


