
Loves Evolution, regulation, cheese and cats. She/Her





www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
1) Variants on both copies of #RNU4-2 cause a recessive neurodevelopmental disorder with prominent speech delay
2) One of the hallmarks is distinct white matter changes on MRI
3) It is clinically and genetically distinct from #ReNU syndrome
www.medrxiv.org/content/10.1...

1) Variants on both copies of #RNU4-2 cause a recessive neurodevelopmental disorder with prominent speech delay
2) One of the hallmarks is distinct white matter changes on MRI
3) It is clinically and genetically distinct from #ReNU syndrome
www.medrxiv.org/content/10.1...
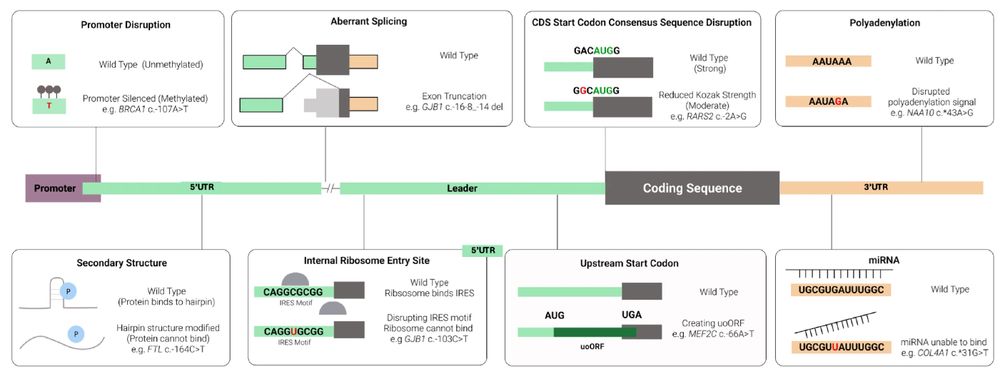
The review outlines how UTR variants cause disease, such as:
- Create or remove upstream AUGs (uAUGs)
- Alter splicing
- Alter polyadenylation
- Interfere with miRNA or protein binding
The review outlines how UTR variants cause disease, such as:
- Create or remove upstream AUGs (uAUGs)
- Alter splicing
- Alter polyadenylation
- Interfere with miRNA or protein binding

Variants that occur outside of protein-coding regions represent a modest but appreciable contribution to rare disease, and should be routinely incorporated into diagnostic pipelines.
(We present a systematic framework for doing this!)
Variants that occur outside of protein-coding regions represent a modest but appreciable contribution to rare disease, and should be routinely incorporated into diagnostic pipelines.
(We present a systematic framework for doing this!)
link.springer.com/article/10.1...

link.springer.com/article/10.1...

