Alex de Mendoza
@alexdemendoza.bsky.social
Evolutionary epigenomics ( eukaryotes / Transcription Factors / Transposable Elements / DNA methylation ) @ QMUL (London).
Lab website: https://www.demendozalab.com/
Lab website: https://www.demendozalab.com/
Then we surveyed species that kept 5mC for giant virus endogenization events, following on our previous work. We find that this is quite common across very distantly related species, rendering support that 5mC can help the host tolerate large influx of potentially deadly DNA. So no more n = 1! 5/7

July 28, 2025 at 10:03 AM
Then we surveyed species that kept 5mC for giant virus endogenization events, following on our previous work. We find that this is quite common across very distantly related species, rendering support that 5mC can help the host tolerate large influx of potentially deadly DNA. So no more n = 1! 5/7
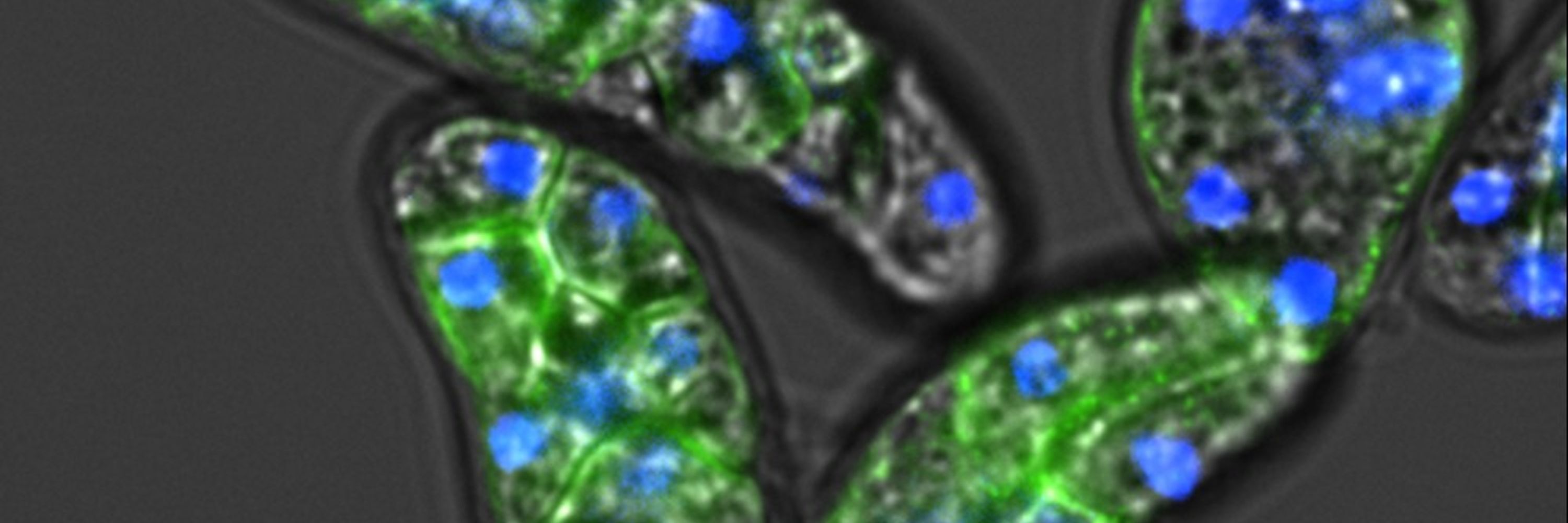
Then we focus on 3 species that kept DNMTs from previously unstudied eukaryotic supergroups : Acanthamoeba, Naegleria & Cyanophora. All show 5mC as a repressive mark, marking transposons and silenced genes together with heterochromatin (H3K9me3 & K27me3), with no hint of gene body methylation. 3/7

July 28, 2025 at 10:03 AM
Then we focus on 3 species that kept DNMTs from previously unstudied eukaryotic supergroups : Acanthamoeba, Naegleria & Cyanophora. All show 5mC as a repressive mark, marking transposons and silenced genes together with heterochromatin (H3K9me3 & K27me3), with no hint of gene body methylation. 3/7
We first characterise the evolution of DNMTs in eukaryotes. Unlike other gene families that expanded from the eukaryotic ancestor, DNMTs come from various prokaryotic sources, integrating throughout eukaryotic history. However, DNMT presence is very patchy, implying pressure for simplification. 2/7

July 28, 2025 at 10:03 AM
We first characterise the evolution of DNMTs in eukaryotes. Unlike other gene families that expanded from the eukaryotic ancestor, DNMTs come from various prokaryotic sources, integrating throughout eukaryotic history. However, DNMT presence is very patchy, implying pressure for simplification. 2/7
So happy to be in 🇭🇰 "de-virtualising" collaborators, talking about TFs and epigenetics, such a welcoming scientific community at HKU. I came for the science, but there's always time for dim sum, dai pai dongs, cha chaan teng and the charm of the old Hong Kong...


April 25, 2025 at 9:24 AM
So happy to be in 🇭🇰 "de-virtualising" collaborators, talking about TFs and epigenetics, such a welcoming scientific community at HKU. I came for the science, but there's always time for dim sum, dai pai dongs, cha chaan teng and the charm of the old Hong Kong...
Not only the 5 nucleobases... but the meteorite contains 5mC. So epigenetics existed even before genetics. Something for you @maxvcg.bsky.social

January 29, 2025 at 6:18 PM
Not only the 5 nucleobases... but the meteorite contains 5mC. So epigenetics existed even before genetics. Something for you @maxvcg.bsky.social
How can these Sox make iPSC? Through partnering with Oct4, and probably also other POU factors. Intriguingly, the same choanoflagellates with SOX genes also posses POU orthologues! However, these are divergent and can't bind DNA as animal POUs do (and can't make iPSC either).5/6

November 14, 2024 at 5:15 PM
How can these Sox make iPSC? Through partnering with Oct4, and probably also other POU factors. Intriguingly, the same choanoflagellates with SOX genes also posses POU orthologues! However, these are divergent and can't bind DNA as animal POUs do (and can't make iPSC either).5/6
Unicellular Sox genes are sister group to the rest of animal Soxes. Yet they already bind the same DNA motifs. So we tested their capacity to substitute an animal Sox, in particular, Sox2 in the classic Yamanaka cocktail. We could generate iPSC, and these could make chimeric "choano"-mice! 3/6

November 14, 2024 at 5:15 PM
Unicellular Sox genes are sister group to the rest of animal Soxes. Yet they already bind the same DNA motifs. So we tested their capacity to substitute an animal Sox, in particular, Sox2 in the classic Yamanaka cocktail. We could generate iPSC, and these could make chimeric "choano"-mice! 3/6
This has been a fully collaborative project with Ralf Jauch group in Hong Kong University, none of this would be possible without their enthusiasm, expertise and effort. It all started with an email about the origins of these genes and a BLAST search. The distribution of hits, was intriguing... 2/6

November 14, 2024 at 5:15 PM
This has been a fully collaborative project with Ralf Jauch group in Hong Kong University, none of this would be possible without their enthusiasm, expertise and effort. It all started with an email about the origins of these genes and a BLAST search. The distribution of hits, was intriguing... 2/6

