
@theacmg.bsky.social #ACMGMtg25 today
👇
Come see me at poster 101 to discuss PMS2, lower penetrance and NCCN guidelines.
And at poster 217 on how we are helping variant interpretation with a resource for the PS4 criteria.

@theacmg.bsky.social #ACMGMtg25 today
👇
Come see me at poster 101 to discuss PMS2, lower penetrance and NCCN guidelines.
And at poster 217 on how we are helping variant interpretation with a resource for the PS4 criteria.
discussing @conehealth.bsky.social GeneConnect program & the Helix Research Network.
He calls every participant with a positive genetic finding.
=> it leads to a very high genetic counseling uptake after ~90%.


discussing @conehealth.bsky.social GeneConnect program & the Helix Research Network.
He calls every participant with a positive genetic finding.
=> it leads to a very high genetic counseling uptake after ~90%.
This study shows that clinical actions were taken for individuals with a FH pathogenic variant + it leads to lower LDL-C.
It also shows some of the care gaps.
Great foundation to increase use of genetics in healthcare. Need more of these.
👏 authors & 8 health systems 👇

This study shows that clinical actions were taken for individuals with a FH pathogenic variant + it leads to lower LDL-C.
It also shows some of the care gaps.
Great foundation to increase use of genetics in healthcare. Need more of these.
👏 authors & 8 health systems 👇
Patients with new/modified therapies had a mean LDL-C reduction of 59mg/dL and 42.0% achieved the hard goal of ≥50% LDL-C reduction or LDL-C ≤70mg/dL.

Patients with new/modified therapies had a mean LDL-C reduction of 59mg/dL and 42.0% achieved the hard goal of ≥50% LDL-C reduction or LDL-C ≤70mg/dL.
Overall, 33% received new/modified lipid-lowering therapy within up to 3 years. Rate was 55% for those with a newly documented FH diagnosis code.
See below the new LDL-lowering agents and therapeutic changes stratified by pre-screening treatment status

Overall, 33% received new/modified lipid-lowering therapy within up to 3 years. Rate was 55% for those with a newly documented FH diagnosis code.
See below the new LDL-lowering agents and therapeutic changes stratified by pre-screening treatment status
Aim: to evaluate changes in clinical management and LDL-C levels among patients we identified with FH.
We studied 453 participants who had enough retrospective and prospective EHR data available.
Among those, 85% (n=384) did not have a prior clinical FH diagnosis.

Aim: to evaluate changes in clinical management and LDL-C levels among patients we identified with FH.
We studied 453 participants who had enough retrospective and prospective EHR data available.
Among those, 85% (n=384) did not have a prior clinical FH diagnosis.
FH is part of the CDC-Tier 1 conditions because it is very high penetrance & actionable
FH is caused by variants in APOB, PCSK9, and LDLR.
Among 211,263 adults participating in The Helix Research Network,
1,020 had FH. That is 1 in 207.
=> more than 1.5 millions in USA with FH.

FH is part of the CDC-Tier 1 conditions because it is very high penetrance & actionable
FH is caused by variants in APOB, PCSK9, and LDLR.
Among 211,263 adults participating in The Helix Research Network,
1,020 had FH. That is 1 in 207.
=> more than 1.5 millions in USA with FH.
Happy to discuss with anyone interested in this topic.

Happy to discuss with anyone interested in this topic.

Taking a 'risk equivalent' approach, women in the 'low risk' group could delay start of mammogram screening by ~5 years.

Taking a 'risk equivalent' approach, women in the 'low risk' group could delay start of mammogram screening by ~5 years.
pLOF vars with AF<0.001% in BARD1, CDH1, MAP3K1, RAD51C,RAD51D and TP53.
We can now risk stratify the entire pop.

pLOF vars with AF<0.001% in BARD1, CDH1, MAP3K1, RAD51C,RAD51D and TP53.
We can now risk stratify the entire pop.
- Path vars in BRCA1/2 or PALB2,ATM,CHEK2 had high risk (pos contrl)
- Coding vars <0.001% AF (many are VUS) have no impact
- only 1 group had a HR > 2: very rare pLOF in the additional set of genes (ex: rare nonsense in TP53)

- Path vars in BRCA1/2 or PALB2,ATM,CHEK2 had high risk (pos contrl)
- Coding vars <0.001% AF (many are VUS) have no impact
- only 1 group had a HR > 2: very rare pLOF in the additional set of genes (ex: rare nonsense in TP53)
(6 after removing the 2 where >90% of the vars are called Pathogenic).
For each, we identified the carriers, and then did a time to event analysis.

(6 after removing the 2 where >90% of the vars are called Pathogenic).
For each, we identified the carriers, and then did a time to event analysis.
It is a fantastic emerging network of health systems in US (& Canada now) with a single research protocol, Exome+ seq for all participants, and nice EHR data.
First experiment was on breast cancer.

It is a fantastic emerging network of health systems in US (& Canada now) with a single research protocol, Exome+ seq for all participants, and nice EHR data.
First experiment was on breast cancer.
Could be VUSs, or VUS high. But in our hands, aggregate effect of these are close to zero. Maybe we can narrow down AND make interpretation automatic. This is key because one goal is to lower overall burden on healthcare enterprise.

Could be VUSs, or VUS high. But in our hands, aggregate effect of these are close to zero. Maybe we can narrow down AND make interpretation automatic. This is key because one goal is to lower overall burden on healthcare enterprise.
We added an intermediate level looking at rare variants of potential risk.
Variants that don't meet Pathogenic criteria. But that are suspicious enough to exclude a person from 'low risk' category.

We added an intermediate level looking at rare variants of potential risk.
Variants that don't meet Pathogenic criteria. But that are suspicious enough to exclude a person from 'low risk' category.
- no guidelines currently
- perception that it is riskier to make the recommendation to not follow standard guidelines and screen less.
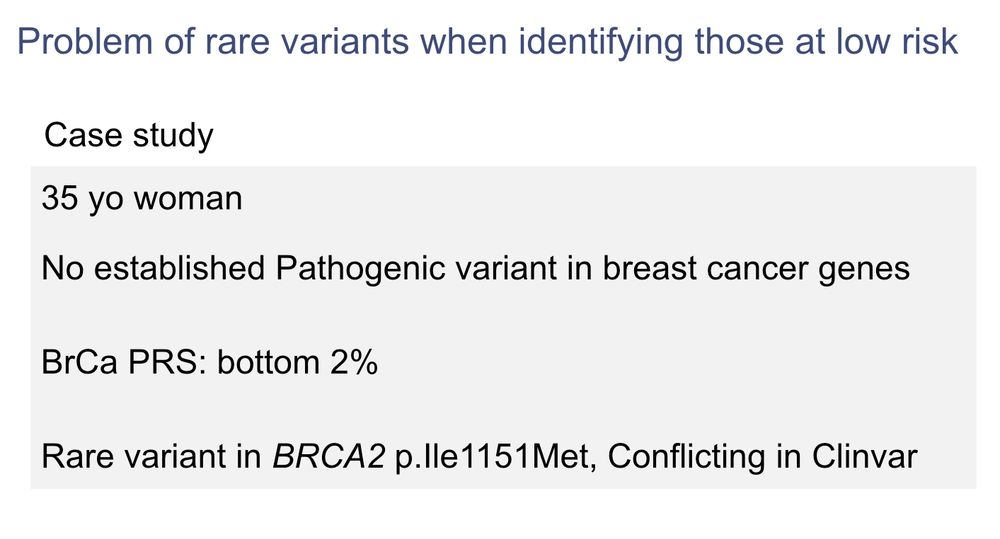
Is it ok to tell the person 👇 she is at low risk of breast cancer ??
- no guidelines currently
- perception that it is riskier to make the recommendation to not follow standard guidelines and screen less.
Is it ok to tell the person 👇 she is at low risk of breast cancer ??
Example of breast cancer 👇

Example of breast cancer 👇
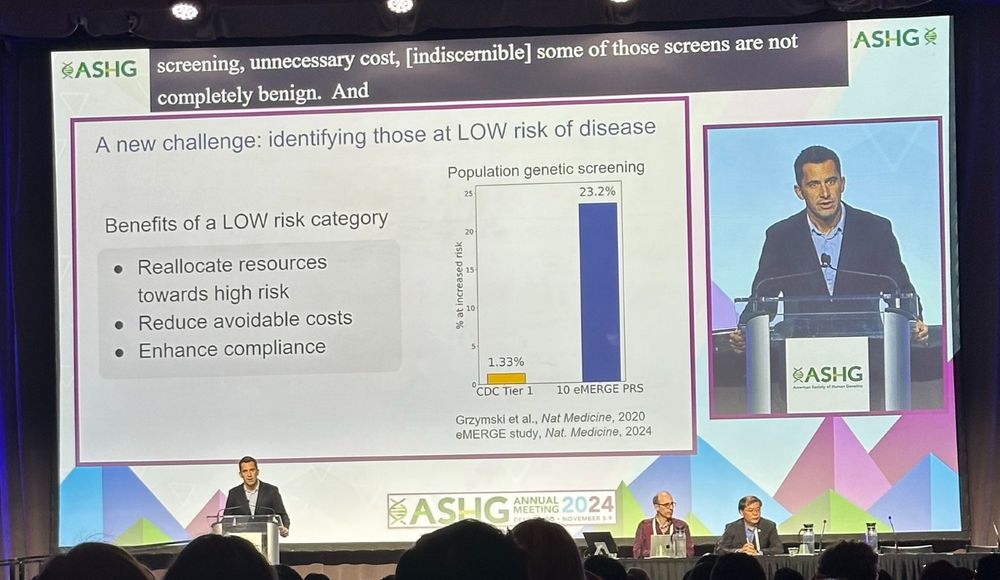
Why a focus on low risk?
- can't only do high risk, high risk, high risk
- clear benefits to patients
- maybe easier to explain avg risk
🧵 with slides
Why a focus on low risk?
- can't only do high risk, high risk, high risk
- clear benefits to patients
- maybe easier to explain avg risk
🧵 with slides

